Edinburgh/DivisionPopper/Design
From 2007.igem.org
Luca.Gerosa (Talk | contribs) |
Luca.Gerosa (Talk | contribs) |
||
| Line 1: | Line 1: | ||
'''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/References|Background]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design]] | [[Edinburgh/DivisionPopper/Realization|Realization]] | [[Edinburgh/DivisionPopper/Modelling|Modelling]] | [[Edinburgh/DivisionPopper/Status|Status]] | [[Edinburgh/DivisionPopper/Conclusions|Conclusions]] | '''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/References|Background]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design]] | [[Edinburgh/DivisionPopper/Realization|Realization]] | [[Edinburgh/DivisionPopper/Modelling|Modelling]] | [[Edinburgh/DivisionPopper/Status|Status]] | [[Edinburgh/DivisionPopper/Conclusions|Conclusions]] | ||
| - | In the Desing | + | In the Desing section we define the architecture (static design) and the mechanisms (dynamic design) of the device. According to Synthetic Biology guidelines, we decouple the design phase from the realization phase. In particular, we work here at the abstraction levels of devices and parts. The parts level is treated only in terms of logical and functional property, thus not going in the detail of actual biological parts. The decision about what biological parts to use and the DNA level details (implementation) is discussed in the [[Edinburgh/DivisionPopper/Realization|Realization]] section. |
| Line 10: | Line 10: | ||
===Device level=== | ===Device level=== | ||
| - | === | + | ===Part level=== |
The Division PoPper functional parts are: | The Division PoPper functional parts are: | ||
| Line 25: | Line 25: | ||
This device outputs a PoPS pulse at each cell division. | This device outputs a PoPS pulse at each cell division. | ||
It can be hooked up to another device such as a counter. | It can be hooked up to another device such as a counter. | ||
| + | |||
| + | ===DNA level=== | ||
==Dynamics Design== | ==Dynamics Design== | ||
Revision as of 21:18, 16 September 2007
MENU : Introduction | Background | Applications | Design | Realization | Modelling | Status | Conclusions
In the Desing section we define the architecture (static design) and the mechanisms (dynamic design) of the device. According to Synthetic Biology guidelines, we decouple the design phase from the realization phase. In particular, we work here at the abstraction levels of devices and parts. The parts level is treated only in terms of logical and functional property, thus not going in the detail of actual biological parts. The decision about what biological parts to use and the DNA level details (implementation) is discussed in the Realization section.
Contents |
Static Design
Device level
Part level
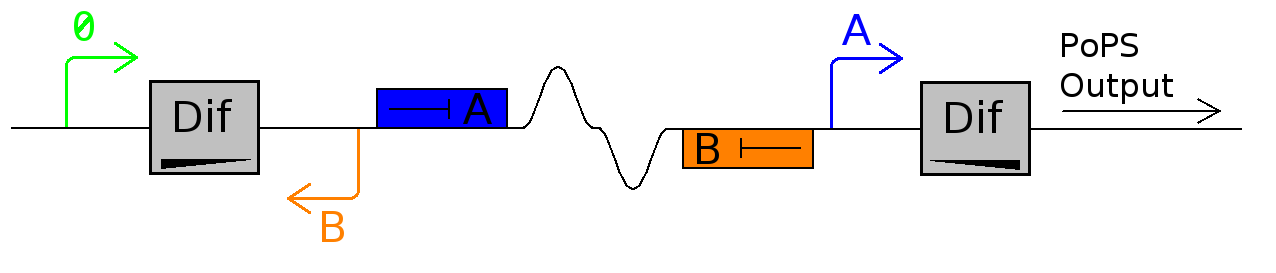
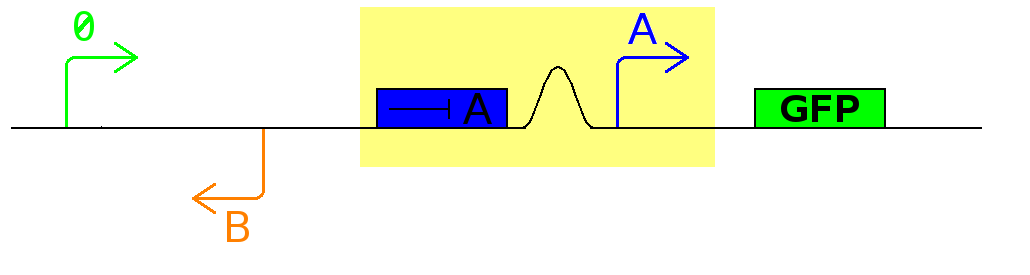
The Division PoPper functional parts are:
- two dif sites sequences.
- a promoter A that can be repressed by protein A.
- a coding region for protein A.
- a promoter B that can repressed by protein B.
- a coding region for protein B.
- a promoter P.
This device outputs a PoPS pulse at each cell division. It can be hooked up to another device such as a counter.
DNA level
Dynamics Design
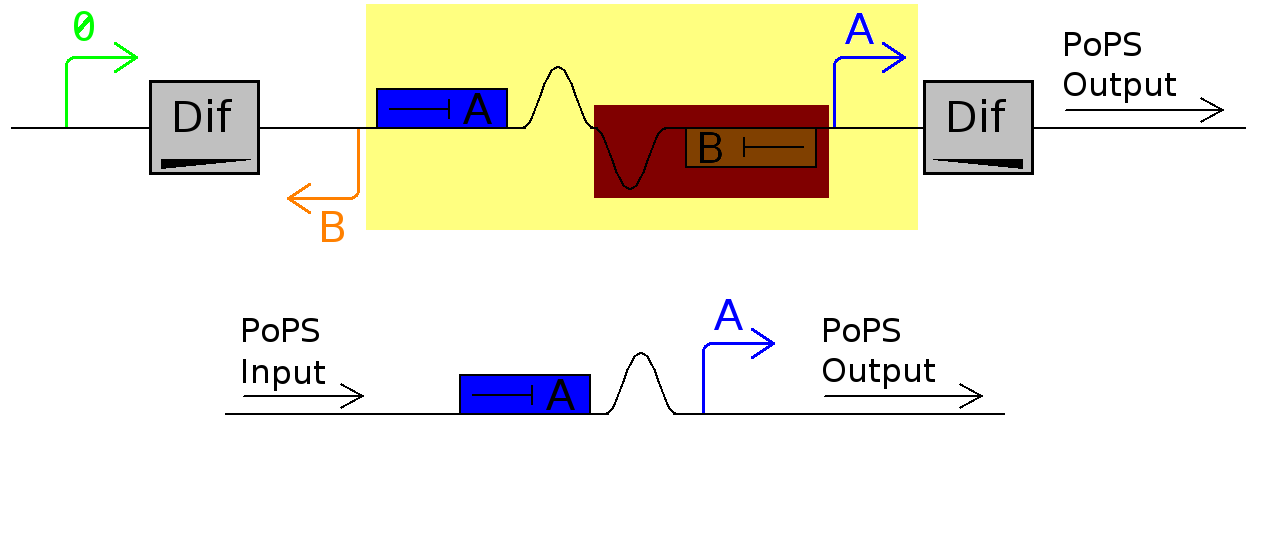
For this device to work, dif site-enclosed DNA needs to flip but once per division. Like all recombination sites, dif-sites are directional and bacteria uses directly repeated dif-sites to resolve genome dimers. Research shows that this resolution occurs only at septation, and we use this temporal control to inverse a plasmid-kept sequence to induce different functions. In its current configuration, Promoter A is repressed by continually produced Repressor A. At this time, no Repressor B is produced.
As cell division occurs, DNA enclosed by two inverted dif sites is reversed. Initially there is no Repressor B present in the cell so Promoter B produces PoPS output. As time passes, Repressor B production is turned on and 'turns off' Promoter B. Meanwhile Repressor A is no longer produced and degrades so that, at the next division, Promoter A can produce PoPS output and the process repeats.
Assumptions
- The Dif sites flip only once during each cell division
- Repressors A and B are produced and degrade faster than the cell cycle
- Promoter B does not interfere with production of repressor A
Proof of Concept
As stated before, the device relies on the dif site flipping only once per cell division. To test whether or not this actually happens, we have devised two simpler experiments.
Exp 1
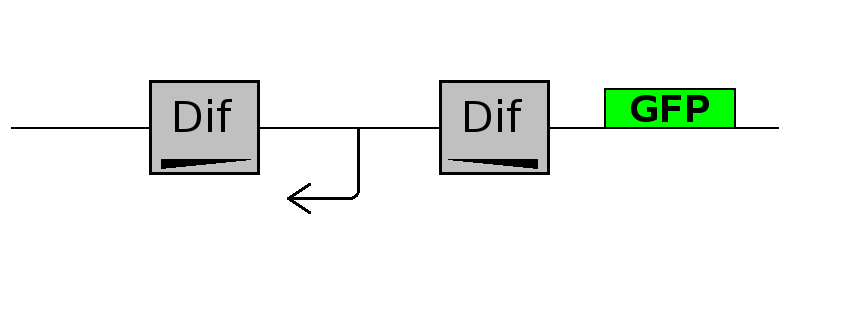
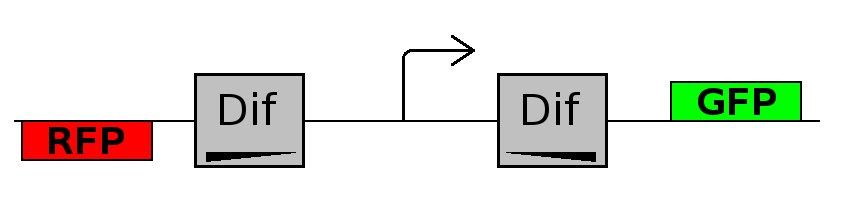
This experiment will prove whether or not the DNA between the two dif sites flip at all during cell division. If the a flip occurs, then the direction the promoter operates will be changed and GFP will be expressed.
Exp 2
This investigates the number of flips that occurs during division and will require rapidly degrading fluorescent proteins.
After each division we expect to see a change in colour as the promoter activates a different reporter.
Exp 3
Currently we are planning to test how the dif sites flip using exps 1 & 2. The final device requires a few more parts, each of which needs to be tested.
We might wish to test the repressors without the dif sites
The repressor part of the system is simply a pair of inverters.
There are 3 basic types of inverter in the registry:
- TetR BBa_Q04400 “this inverter functions well. [jb, 5/24/04]”
- CI (Lambda) BBa_Q04510 “this inverter functions well. [jb, 5/24/04]”
- LacI BBa_Q04121 "a strong 'on' state with significant background in the 'off' state. [jb,5/24/04]”
exp 3 design
This will test to see if promoter B causes problems with promoter 0's promoting repressor A and tests the standard repressor inverter found in the registry.
Parts required:
- Promoter 0 – suggest BBa_I0500 (arabinose) – Plate 2, 9I
- Promoter B – reverse complimentary lacI promoter – Used in exp 1
- Inverter – suggested BBa_Q04510 (CI (Lambda)) – Plate 2, 13K
- Reporter – use same as experiment 1
Biobricks Required
Biobricks From Registry
| Part | For use in | Brick used |
|---|---|---|
| Reporter | Exp1, Exp2, Exp3 | [http://partsregistry.org/Part:BBa_E0422 BBa_E0422] - Fast Degrading ECFP |
| Inverter | Exp3 | [http://partsregistry.org/Part:BBa_Q04510 BBa_Q04510] - CI (Lambda) Inverter |
| Promoter 0 | Exp3, Final PoPper | |
| Promoter A | Final PoPper | |
| Repressor (+RBS) A | Final PoPper | |
| Central Terminator | Final PoPper |
Biobricks requiring construction
| Part | For use in |
|---|---|
| Dif sites | Exp1, Exp2, Final PoPper |
| Reversed Promoter B | Exp1, Exp3, Final PoPper |
| Reversed Repressor (+RBS)B | Final PoPper |