Edinburgh/Yoghurt/Design
From 2007.igem.org
(→Gene Expression In Lactobacillus) |
|||
| Line 76: | Line 76: | ||
[[Image:Multihost plasmid.jpg|thumb|200 px]] | [[Image:Multihost plasmid.jpg|thumb|200 px]] | ||
| + | In order to express our flavour and colour genes in lactic acid bactia (LAB), we require a different vector to those currently in use by the registry. After some research, we found that a few groups withint the UK were working with LAB. | ||
| + | We kindly thank Dr Mike Gasson & Dr. Claire Shearman, of the Institute of Food Research in Norwich for generously donating the pTG262 vector for our iGEM2007 project. | ||
| + | '''pTG262''' | ||
| + | |||
| + | This vector was ideal for use as a biobrick chassey, as: | ||
| + | * it contained three of the four biobrick restriction enzyme sites (insertion of a biobrick added the other two) | ||
| + | * it has a gram positive origin of replication, which works well in ''E. coli'' (enabling the plasmid to act as a shuttle vector between ''E. coli'' and LAB | ||
| + | * contains three antibiotic resistance sites (chloramphenicol, neomycin & gentamicin) | ||
| + | * known to work in ''Lactobacillus'', ''Lactococcus'' and ''Bacillus'' as well as ''E. coli'' | ||
| + | |||
| + | We also plan to test the efficieny of the vector in other gram negative bacteria including; ''Shewanella'' & ''Pseudomonas'', ''Agrobacterium'' | ||
---- | ---- | ||
[[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Objectives|Objectives]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/References|References]] | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Objectives|Objectives]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/References|References]] | ||
---- | ---- | ||
Revision as of 13:08, 1 October 2007
 Introduction | Applications | Objectives | Design | Modelling | Wet Lab | References
Introduction | Applications | Objectives | Design | Modelling | Wet Lab | References
Contents |
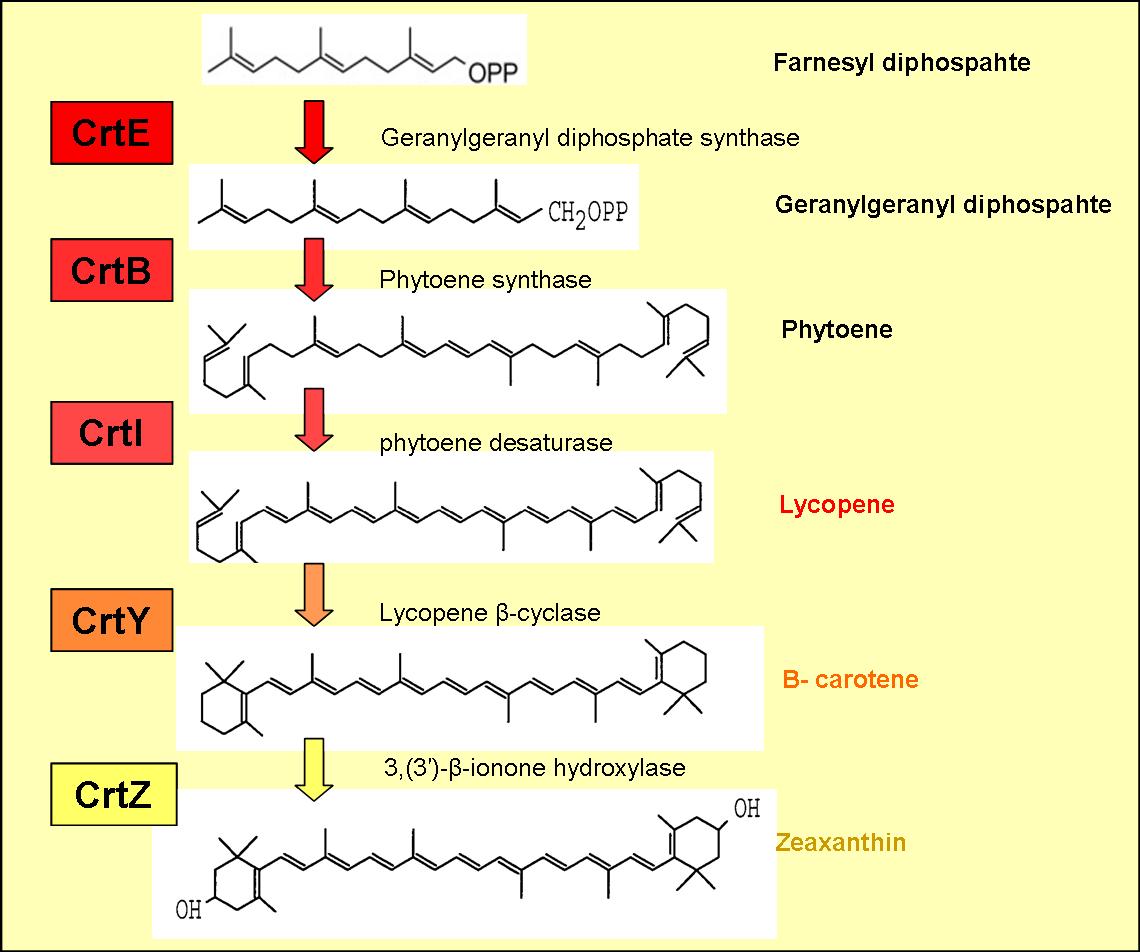
Colour Production
The zeaxanthin operon is naturally found in many plants and bacteria. The operon encodes five genes, which enable the production of the yellow pigment zeaxantin.
The five genes present in the zeaxanthin operon are:
- CrtE
- CrtB
- CrtI
- CrtY
- CrtZ
The enzymes encoded by these genes and the products formed are displayed in figure 1.
There are three pigments produced by the genes with in the zeaxanthin operon and a brief description of each is given below:
Lycopene
The genes CrtE, CrtB and CrtI produce lycopene from farnesyl diphosphate (an intermediate in the mevlonate pathway).
Lycopene is a red pigment (found in tomatoes), which also has extremally powerful antioxidant properties and may possibly help protect you from cancer.
B-carotene
B-carotene is produced by cyclising lycopene, which is carried out by lycopene B-cyclase encoded by the gene CrtY (see figure )
B-carotene has an orange pigmentation and is responsible for the colour of carrots, winter squash and several other vegetables. The pigment can be stored in the liver and converted to Vitamin A, a form of retinol, required for sight.
zeaxanthin
Addition of CrtZ to the CrtEBIY construct enables the hydroxylation of B-carotene (see figure 1) to zeaxanthin, which is a yellow pigment.
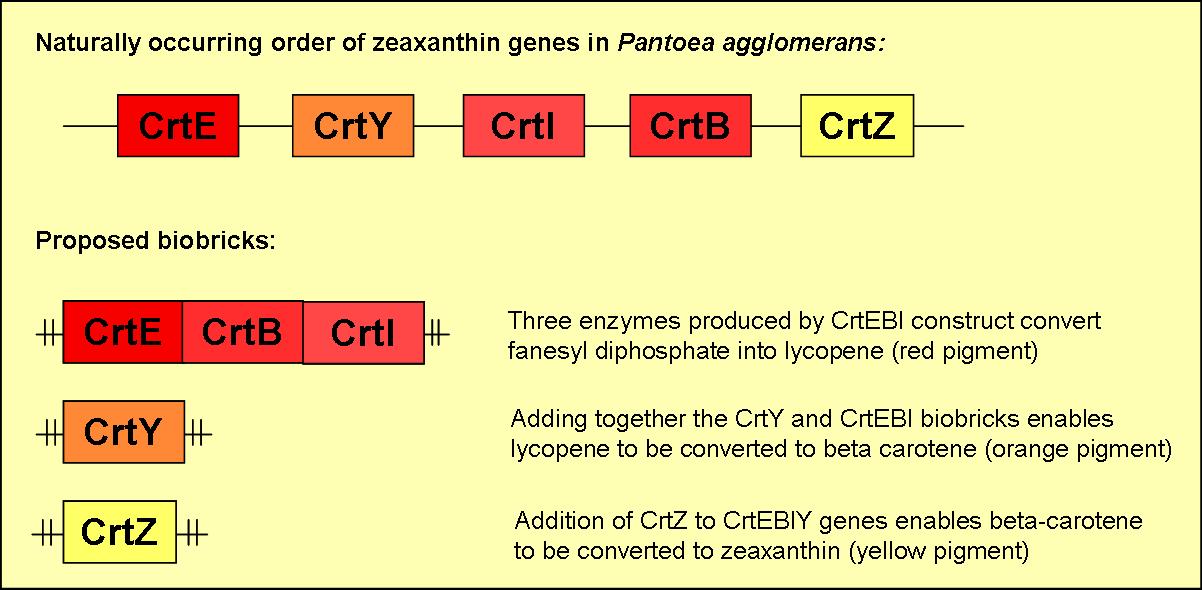
Proposed Biobricks
We plan to produce five different biobricks with varying combinations of the five zeaxanthin genes. A brief overview of three of these biobricks can be found in figure 2.
Biobrick 1
- will consist of the three genes CrtE, CrtB & CrtI
- will produce the red pigment lycopene
Biobrick 2
- will contain CrtY
- when induced in the presence of functional CrtEBI genes an orange pigment (B-carotene) will be formed
Biobrick 3
- will consist of CrtZ
- when induced in the presence of functional CrtEBIY genes it will enable the formation of the yellow pigment zeaxanthin
Biobrick 4
- will contain the four genes CrtE, CrtB, CrtI & CrtY
- the construct will be capable of producing B-carotene
Biobrick 5
- will contain all five genes present in the zeaxanthin operon; CrtE, CrtB, CrtI, CrtY & CrtZ
- biobrick will enable the production of zeaxanthin (yellow pigmentation)
Vanilla Flavour Production
Lemon Flavour Production
Gene Expression In Lactobacillus
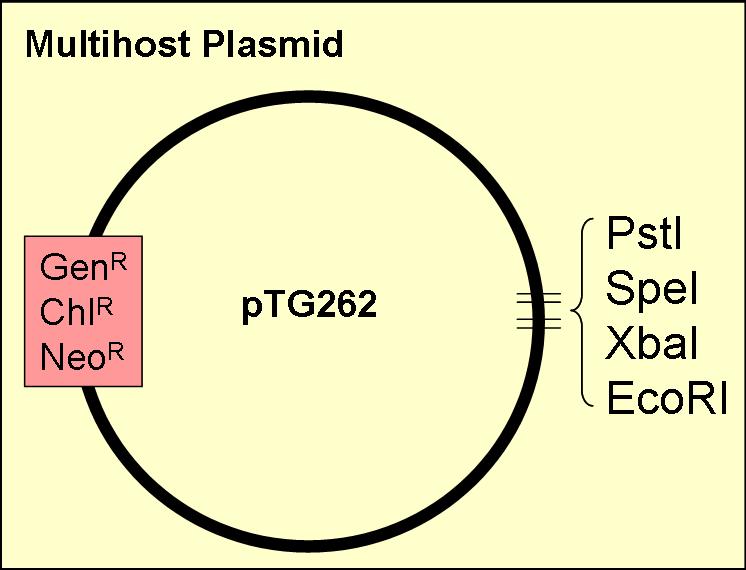
In order to express our flavour and colour genes in lactic acid bactia (LAB), we require a different vector to those currently in use by the registry. After some research, we found that a few groups withint the UK were working with LAB.
We kindly thank Dr Mike Gasson & Dr. Claire Shearman, of the Institute of Food Research in Norwich for generously donating the pTG262 vector for our iGEM2007 project.
pTG262
This vector was ideal for use as a biobrick chassey, as:
- it contained three of the four biobrick restriction enzyme sites (insertion of a biobrick added the other two)
- it has a gram positive origin of replication, which works well in E. coli (enabling the plasmid to act as a shuttle vector between E. coli and LAB
- contains three antibiotic resistance sites (chloramphenicol, neomycin & gentamicin)
- known to work in Lactobacillus, Lactococcus and Bacillus as well as E. coli
We also plan to test the efficieny of the vector in other gram negative bacteria including; Shewanella & Pseudomonas, Agrobacterium
Introduction | Applications | Objectives | Design | Modelling | Wet Lab | References