Edinburgh/Yoghurt/Wet Lab
From 2007.igem.org
 Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | References
Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | References
Contents |
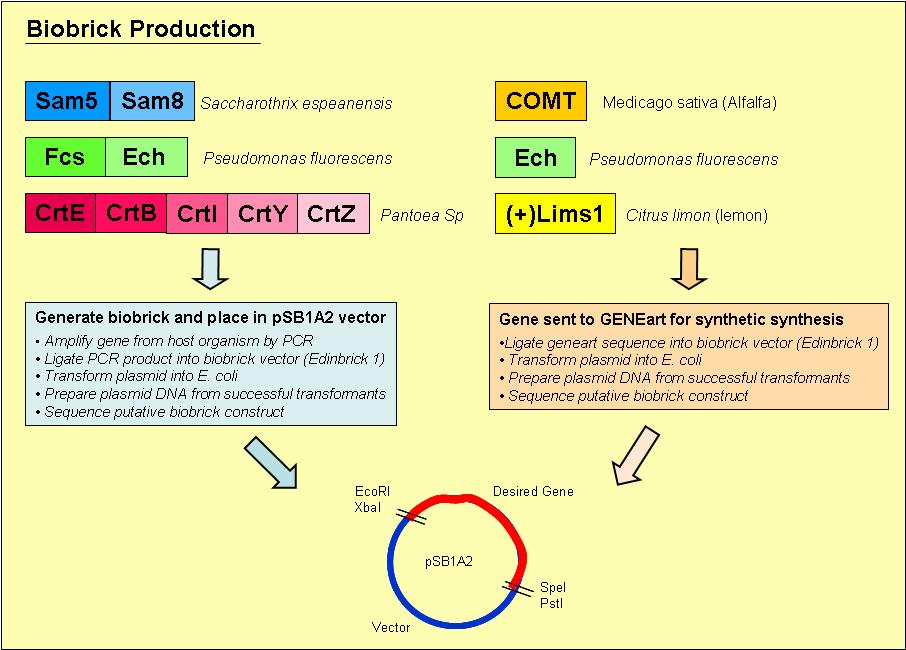
Biobrick Creation
Overview of the process we used to generate our biobricks:
1. Designed primers, which included a ribosome binding site and the biobrick restriction sites, EcoRI, XbaI, PstI and SpeI, to amplify our desired gene out of its host organism
2. Purified the PCR product and loaded onto an agrose gel to check for the presence of PCR product of the right size
3. If a band was present at the right size, we digested the PCR product and vector with the restriction enzymes EcoRI & PstI
4. Digested PCR product and vector were ligated overnight with T4 DNA ligase
5. Ligated vector and gene were then transformed into E. coli and plated onto Blue/ White selection Amplicillin plates
6. Transformants that contained a product ligated into the vector would grow as white colonies and are easy to select
7. Minipreps were prepared of the white colonies
8. Purified vector DNA was digested with restriction enzymes EcoRI and PstI to determine if the vector insert was of the correct size
9. Vectors containing inserts of the correct size were sent for sequencing to confirm if they did indeed contain the correct gene
The complete methods we used to generate our biobricks may be found on the French Lab OpenWetWare site
Zeaxanthin Synthesis Pathway
Biobricks created so far
- CrtE
- CrtBI (with PstI restriction sites)
- CrtZ
Primers
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| CrtE | crtE f1: gca gagctc gcgt tgcc gtaa atgt atc | crtE r1: aa actagt gcga tcgc cgcg aaat g |
| CrtBI | crtI f1: tga gagctc atcg ttaa agag cgac | crtB r1: gc actagt caaa actt cagg cgac |
| CrtY | crtY f1: cag gagctc ttaa gtgg gagc ggct atg | crtY r1: ac actagt tggt ttca tgta gtcg |
| CrtZ | crtZ f1: aga gagctc tacc ggag aaat tatg | crtZ r1: cc actagt cagg ccct tact tccc |
More information on the primers used to amplify the zeaxanthin pathway genes may be found on the French Lab OpenWetWare site
Lycopene Biobrick
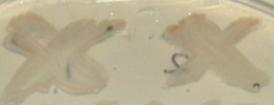
Success at last!
We have managed to successfully clone CrtE and CrtIB genes and ligate them together in a biobrick, which produced lycopene.
Figure 2, shows some E. coli colonies, which have produced a small amount of lycopene and have started to turn red.
We have made two different variations of the lycopene biobrick, one which still included the two forbidden restriction sites in CrtI, and one where the two restriction sites have been mutated out.
B-carotene Biobrick
B-carotene biobrick in registry
Zeaxanthinin Biobrick
Vanillin Biosynthesis Pathway
Biobricks created so far
- Sam8
- Sam5
- LacZ/Sam8/Sam5 three gene construct
Genes sent to GENEART for synthesis
- COMT
- ech
Primers
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Sam5 | sam5 f1: at gaattc gcggccgc t tctag atg acc atc acg tca cct g | sam5 r1: ct actagt a tta tta ggt gcc ggg gtt gat cag |
| Sam8 | sam8 f1: at gaattc gcggccgc t tctag atg acg cag gtc gtg gaa cg | sam8 r1: ct actagt a tta tta tcc gaa atc ctt ccc gtc |
| ech | ech f1: aat gaattc gcggccgc t tctag atg agc aaa tac gaa ggt c
ech f2: atc gagctc acacc cagaa caaga gc | ech r1: ct actagt a tta tta gcg ttt ata ggc ttg cag c
ech r2: tt actagt atcgg gaaca cgttc aagc |
| fcs | fcs f1: aat gaattc gcggccgc t tctag atg cgc tcc ctg gaa ccc
fcs f2: gtg gagctc actga agaac agggc gtg | fcs r1: ct actagt a tta tta cgg ttt ggg ccc ggc ac
fcs r2: aa actagt atgcc gtgac agcaa atagg |
More information on the primers used to amplify the vanillin pathway genes may be found on the French Lab OpenWetWare site
Cloning of Ech and Fcs genes from Pseudomonas flourescens
We decided to clone ech and fcs by directly amplifying the genes up from the P. fluorescens genome. In order to do this we ordered two primer sets (one for each gene), which also encoded the four biobrick restriction sites. In addition the ech reverse primer was modified to point mutate out the PstI restriction site found in the genes 3' end.
After trying many different annealing temperatures, we found that the two genes were not being amplified up by PCR. This could be due to the high GC nature of the P. fluorescens genome. Therefore we decided to modify the PCR reaction with glycerol or DSMO, which are both reported to improve PCR amplification of GC rich organisms. In addition we also designed a second primer set, by searching for sites further away from the gene with a lower GC content, which hopefully would improve primer binding. The second primer sets also contained a ribosome binding site.
The use of the second primer set and glycerol resulted in a PCR product for the fcs gene of ~2kb. This fcs PCR product was then purified, digested with SacI/ SpeI restriction enzymes and ligated into the Edinbrick1 vector. So far we have not yet managed to produce any fcs clones with a viable fcs insert.
Unfortunately there was no viable PCR product for the ech gene. Therefore we decided to send a modified ech gene to GENEART for synthesis. We modified the ech gene by altering the coding reading frame to have the optimum codons for E. coli, this was both to decrease the gc content of the gene and to ensure optimal expression of ech.
Cloning of Sam5 and Sam8 genes from Saccharothrix espeanensis
Just like with the ech and fcs genes we designed primers to amplify up Sam5 and Sam8 from the S. espeanensis genome. Like the ech and fcs genes, we also found that a simple PCR resulted in no gene product, due to S. espeanensis also having a high GC genome (recognising a pattern here).
However glycerol and a higher melting temperature came to our rescue, and by encorporating these two factors into our PCR, we got products for both Sam5 and Sam8.
The PCR amplifications of the two genes were digested with SacI, SpeI restriction enzymes and ligated into the Edinbrick1 vector. Colonies containing either the Sam5 or Sam8 gene were easily identified and minipreps were made of their vector DNA.
The purified vector DNA was then sequenced to confirm the genes present were indeed Sam5 and Sam8. Fortunately for us the sequencing results confirmed we had isolated the right genes.
We then made a Plac-Sam8 construct, to induce Sam8 expression upon the addition of lactose. The Sam5 gene was also ligated into this construct.
The next stage is to test the Plac-Sam8-Sam5 construct, to determine if native proteins are synthesised and the correct enzymatic reactions are carried out. We plan to do this by inducing tyrosine ammonia lyase (sam8) and p-coumoryl-4-hydroxylase (sam5) by growing E. coli on lactose media containing tyrosine.
Gas chromatography coupled with mass spectrometry (GC-MS) will then be used to test for the production of p-coumaric acid and caffeic acid.
Cloning of COMT from Alfalfa (Medicago sativa)
Database searches revealed that the only COMT genes from Alfalfa which had been sequenced were all cDNAs, thus we had no idea as to whether the native COMT gene contained introns or not. Therefore we decided to both amplify the gene up from the Alfalfa plant, and to obtain a cDNA clone via reverse transcription.
The PCR amplification of COMT straight from the Alfalfa gene revealed there was no gene product present of ~1.1kb in size. From this we concluded the COMT gene must contain introns and proceeded to try and clone the gene via reverse transcription of COMT mature mRNA. Unfortunately for us we experienced some difficulty with producing COMT cDNA and decided to have COMT synthesised by GENEART.
Multi Host Plasmid pTG262
The plasmid we recieved had a multicloning site containing EcoRI, XbaI and PstI sites. To convert pTG262 into a biobrick vector we inserted a biobrick between the EcoRI and PstI sites, this simultaneously removed the intravening XbaI site, and introduced all four biobrick restriction enzymes sites (EcoRI, XbaI, SpeI & PstI).
To create the biobrick restriction sites, we inserted a total of three biobricks:
- Plac-RFP (gives red transformants)
- Plac-lacZ (gives blue transformants)
- Ptet-RFP
In order to test the plasmid in a multitude of gram negative bacteria, including Shewanella, Pseudomonas & Agrobacterium, we will transform the pTG262-Plac/RFP construct into our chosen host. This will enable easy detection, as all bacteria capable of retaining and expressing the vector will form easily detected red colonies, when grown on media containing lactose.
Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | References