Paris/A synthetic multicellular organism
From 2007.igem.org
Contents |
Why a synthetic multicellular organism ?
A major challenge for synthetic biology will be to achieve complexity. It will not be trivial to implement complex systems within single cells, especially given the fact that most of the devices built until now contain the same biobricks (ptet, pLac, pLambda, pBAD LuxI, AHL...). An answer to this problem may be multicellularity. If you need, for instance, a switch and an oscillator, but the ones that would suit are made out of the same basic components, then you're going to have a problem! Unless you are able to implement them in two different cell lines of a synthetic multicellular bacteria !
An other point of view, is that building a synthetic multicellular organism out of unicellular bacteria may help shedding light upon the emergence of multicellularity. What type of selective pressure is liable to drive evolution towards multicellular organisms? What kind of mechanisms are needed to achieve multicellularity? ...
How can we define a multicellular organism ?
There is no universally admitted definition. Wikipedia gives: "an organism (in Greek organon = instrument) is a living complex adaptive system of organs that influence each other in such a way that they function in some way as a stable whole."
For us, the organs will simply be differentiated cells. The definition we retain is that of a multicellular entity with different types of cells fulfilling different complementary tasks.
In multicellular organisms, some cells differentiate to realize a function useful for the whole organism. Useful means that it will give a better fitness to the organism, or in other terms, that it will help producing more offspring. The specialized cells are usually not the same than the ones reproducing the organism, and they may even lose this ability. Thus, there is a notion of sacrifice of some cells to the profit of those dedicated to reproduction. The cells able to reproduce the organism represent the germline, the ones which specialize and lose this ability represent the soma.
How to proceed ?
Our aim is to make E.Coli differentiate into two distinct lines. Each line depending on the other one, the survival of both is necessary. The relation between our cell lines is inspired of the classic soma/germline specialization of multicellular organisms. One of our lines (the germline) will be able to reproduce the organism, but to do so, it will need the presence of the other line (the soma) which itself is unable to reproduce. The germline will produce the soma through differentiation of part of its cells. Since it is unable to reproduce, the soma has no existence without the germline, and the germline needs the soma to reproduce. There is thus an interdependence relationship.
The way we choose the germline to be dependent on the soma is inspired of crossfeeding experiments. It will be auxotroph for a given nutriment. This nutriment will be provided by the soma.
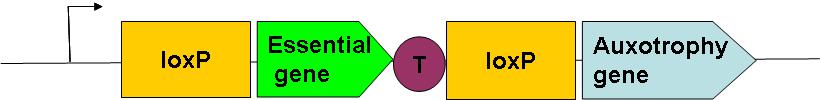
How can we drive a cell differentiation into two distinct lines? The differentiation can either be genetic or epigenetic. In epigenetic differentiation, the two or more lines are distinguished only by different patterns of protein expression, generated by an appropriate biochemical circuit. In genetic differentiation however, individual lines will end up having different genome sequences. As a mean of implementing differentiation, we have chosen the genetic solution based on a DNA recombination system. We will introduce a special genetic construct into the bacterial chromosome; in a subset of cells in a population the excision of the genomic cassette will lead to differentiation of these cells into soma. The construct could be simply represented in the following way:
This is the construct of the germline. Here, the essential gene is expressed, but the auxotrophy gene is not, transcription being stopped at the termination site represented by the red circle with T. Thus this strain is auxotroph for the metabolite this "auxotrophy gene" would produce if expressed. If the CRE recombinase is expressed, the DNA sequence between the two loxP sites will be excised and so the essential gene leading to the following construction in the daugther cell:
Here the auxotrophy gene is expressed (the loxP site does not affect the transcription), but the strain doesn't express the essential gene anymore, it is no more there! Such cells will thus die after a given time. But in the meanwhile, it will hopefully have produced enough of the auxotrophy metabolite to feed the germline!
What essential gene ?
The essential gene must fulfill two main criteria:
- The cells should be able to live as long as possible without it.
- Its deletion should not impair too much the capacity of the cell to produce the auxotrophy metabolite
We have chosen the cell division gene FtsK. For more details on our choice, see here.
What auxotrophy gene ?
The auxotrophy gene must fulfill three main criteria:
- The soma cells must be able to overproduce and excrete it
- Their must be a simple auxotrophy to the metabolite
- It would be better if there were a promoter sensitive to the metabolite concentration. If this is the case, we could place the CRE recombinase under the control of this promoter. In this way, the germline will differentiate into soma only if there is a need for the auxotrophy metabolite.
We choose DAP (diaminopimulate). For more details on our choice, see here.
Preliminary work
To convince ourselves and others that our system might actually work, we need mainly two pieces of evidence:
- The soma needs to be able to feed the germline.
- The germline cells need to be able to differentiate into somatic cells at a frequency high enough so that enough somatic cells will be produced to feed the germline, and low enough so that not all the germline will differentiate.
Those two points constituted our first two main axes of research. To see the related experiments click on the links.