Virginia Tech/lab
From 2007.igem.org
BlairLyons (Talk | contribs) m |
BlairLyons (Talk | contribs) m |
||
| Line 72: | Line 72: | ||
| style="padding: 20px; background-color: #FFFFFF;" | | | style="padding: 20px; background-color: #FFFFFF;" | | ||
| + | <h3>Our wet lab work falls into three categories...</h3> engineering a strain of bacteria carrying the reporter plasmid, preparing phage lysates and getting practice working with λ phage, and putting the two together to generate experimental data for our model. | ||
| + | [[Image:Plasmid_vector.PNG|thumb|200px|right|A preliminary plasmid design.]] | ||
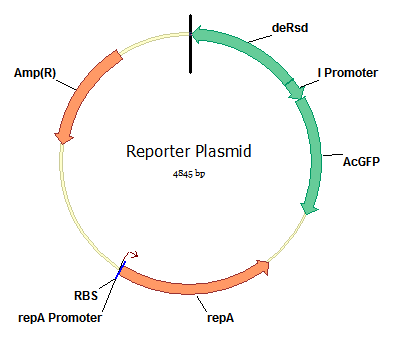
| + | '''Designing and Building the Reporter Plasmid''' | ||
| + | We designed our reporter plasmid using parts of the λ switch and two fluorescent reporter protein genes. We ordered a two-way promoter part modeled off the right promoter of the λ switch, and attempted to use standard BioBrick construction techniques to put it into a plasmid with two fluorescent genes from the registry, GFP and YFP. | ||
| + | '''Read more''' about our [[Virginia_Tech/plasmid_design|reporter plasmid design]] or Virginia_Tech/plasmid_construct|reporter plasmid construction]]. | ||
| + | |||
| + | |||
| + | '''Working with λ Phage''' | ||
| + | |||
| + | We received our phage early in the summer and started using it to infect bacteria. We practiced ways of creating lytic cultures, called lysates, and experimented with ways to take advantage of the fluorescent phage we had received. | ||
| + | |||
| + | '''Read more''' about the [[Virginia_Tech/phage|phage]] and our work with [[Virginia_Tech/phage_lysate|phage lysates]]. | ||
| + | |||
| + | |||
| + | '''Infecting ''E. coli''''' | ||
| + | |||
| + | By infecting ''E. coli'' with λ phage, we gathered data about infection trajectories and were able to observe lysis under a microscope. we gathered trajectory data on a plate reader and then compared it with and matched it to simulated trajectories from our computer model. | ||
| + | |||
| + | Read more about [[Virginia_Tech/infection_model|gathering and matching infection trajectories]] to our model and our [[Virginia_Tech/observe_lysis|observations of lysis]]. | ||
|}<html></center></html> | |}<html></center></html> | ||
Revision as of 00:22, 25 October 2007
|
|
Our wet lab work falls into three categories...engineering a strain of bacteria carrying the reporter plasmid, preparing phage lysates and getting practice working with λ phage, and putting the two together to generate experimental data for our model.
Designing and Building the Reporter Plasmid We designed our reporter plasmid using parts of the λ switch and two fluorescent reporter protein genes. We ordered a two-way promoter part modeled off the right promoter of the λ switch, and attempted to use standard BioBrick construction techniques to put it into a plasmid with two fluorescent genes from the registry, GFP and YFP. Read more about our reporter plasmid design or Virginia_Tech/plasmid_construct|reporter plasmid construction]].
We received our phage early in the summer and started using it to infect bacteria. We practiced ways of creating lytic cultures, called lysates, and experimented with ways to take advantage of the fluorescent phage we had received. Read more about the phage and our work with phage lysates.
By infecting E. coli with λ phage, we gathered data about infection trajectories and were able to observe lysis under a microscope. we gathered trajectory data on a plate reader and then compared it with and matched it to simulated trajectories from our computer model. Read more about gathering and matching infection trajectories to our model and our observations of lysis.
|