Virginia Tech/multi model
From 2007.igem.org
BlairLyons (Talk | contribs) m |
BlairLyons (Talk | contribs) m |
||
| Line 73: | Line 73: | ||
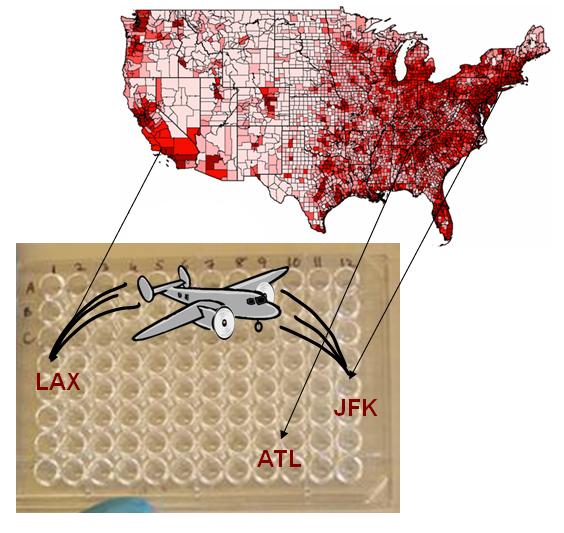
| + | [[Image:Level3.JPG|right|thumb|400px|An illustration of mapping an air traffic network to a 96-well plate.]] | ||
| + | <h3>Tying in Air Travel: Level 3 of the Model</h3> | ||
| + | Our original inspiration for this project was modeling of epidemic spread in the age of air travel. In order to integrate this into our model, we planned a third level of the multi-scale model: infection spread among populations. In order to gather experimental data, we planned to use an automated liquid handling system with various mixing patterns, including a network based on air travel data, to mix aliquots of bacterial culture, both infected and uninfected, around the plate. We hoped to find parameters for which the infection died out and parameters for which it became an epidemic. From our single population experiments with phage, we discovered that λ is so stable that it almost never dies out. If we continue this project, we would engineer a new strain of λ phage that is not so stable, so we could observe the effect of parameters on outcome. | ||
| + | |||
| + | We did not finish this level in the short amount of time we had this summer, but we did obtain air traffic data and start mapping it to our 96-well plate. We also designed our model's [[Virginia_Tech/toolkit|toolkit]] around our automated liquid handling system and its Excel spreadsheet of mixing patterns. If we continued this project, these experiments would be very interesting to complete. | ||
Revision as of 23:05, 25 October 2007
|
|
|
Tying in Air Travel: Level 3 of the ModelOur original inspiration for this project was modeling of epidemic spread in the age of air travel. In order to integrate this into our model, we planned a third level of the multi-scale model: infection spread among populations. In order to gather experimental data, we planned to use an automated liquid handling system with various mixing patterns, including a network based on air travel data, to mix aliquots of bacterial culture, both infected and uninfected, around the plate. We hoped to find parameters for which the infection died out and parameters for which it became an epidemic. From our single population experiments with phage, we discovered that λ is so stable that it almost never dies out. If we continue this project, we would engineer a new strain of λ phage that is not so stable, so we could observe the effect of parameters on outcome. We did not finish this level in the short amount of time we had this summer, but we did obtain air traffic data and start mapping it to our 96-well plate. We also designed our model's toolkit around our automated liquid handling system and its Excel spreadsheet of mixing patterns. If we continued this project, these experiments would be very interesting to complete.
|