Cambridge/Amplifier project
From 2007.igem.org
Contents |
Introduction
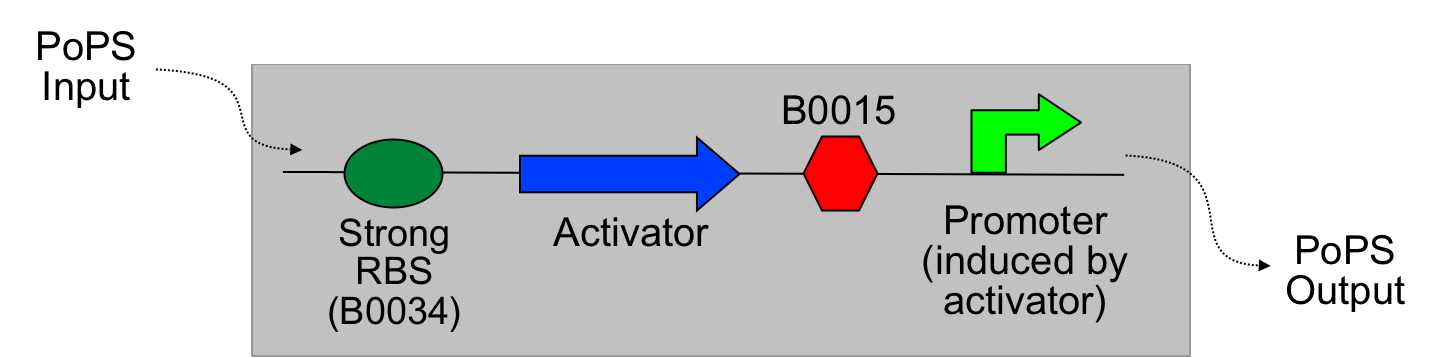
The main idea of this part of the project was to build a synthetic amplifier which would take a PoPS input and would give a PoPS output at a higher level:
A PoPS input is used to generate the activator protein which in turn activates the promoter to lead to an (amplified) PoPS output.
nb: PoPS is the flow of RNA polymerase molecules along DNA (i.e., 'current' for gene expression). The PoPS level is set by the amount of RNA polymerase molecules that pass a specific position on DNA each second.
Purpose of amplifier
Signalling is an integral part of all systems; it enables control and communication to exist such that coordinated responses can be formed. Signals can vary in their strength and are often difficult to detect, especially with regards to low copy number plasmids or small population sizes. We sought to develop a set of well characterised synthetic amplifiers which could be used as standalone devices to increase the strength of a signal by a known factor. The benefits of such a synthetic device would be to improve detection techniques enabling easier testing of systems due to the reduced minimum expression rate. The synthetic amplifier could also be used to alter the characteristics of natural occurring systems by altering their sensitivity.
Many synthetic signalling systems are formed of molecules which naturally occur within the bacteria in which they are being utilised. This can cause many complications due to cross talk which interferes with the cellular activities. We desired the synthetic amplifiers to use signalling that was not commonly used within the natural life-cycle of the bacteria. This would enable them to be used as devices to aid development within synthetic biology without the requirement to have a detailed understanding of the underlying workings of the device, thus making them easily usable and functional.
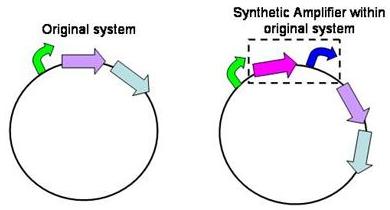
Structure of the amplifier
The amplifiers work by having a gene (the activator) transcribed downstream of the original system promoter which acts on a specific ‘amplifier’ promoter upstream of the area to be amplified.
The activator would be transcribed at the rate dictated by the original systems promoter; it would then be translated and bind to the ‘amplifier’ promoter which would then dictate the rate of transcription of the rest of the system. Given the ‘amplifier’ promoter having a higher rate of transcription than the original system promoter there would be an up regulation of transcription and therefore amplification.
For the amplifiers we have used phage activator polypeptides that are known to activate transcription from phage promoters:
Bacteriophage PSP3 and fR73 Activator Proteins: Analysis of Promoter Specificities; Julien and Calendar, 1996; JOURNAL OF BACTERIOLOGY, Oct. 1996, p. 5668–5675 (link: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=178405)
This paper characterises the interactions of four transcriptional activators and six promoters.:
The promoters are the PF, PO, PP and PV promoter from P2 phage and the Psid and PLL promoters from its satellite phage P4.
They are activated by a family of activators, four of which are: P2 ogr, PSP3 pag, phiR73 delta, and P4 delta.
We chose to use the activators and promoters detailed within the above paper because they are bacteriophage components so are not common in cellular interaction. This achieves our goal of having synthetic amplifiers which will not conflict with normal cellular activity. There is also a nice set of possible constucts as each of the activator genes acts on all of the promoters, hence the possibility of developing a family of constructs to be used as amplifiers. The above paper also showed different induction levels of the same promoter when activated by different polypeptide activators and a difference in activational strength of the activators on different promoters. This gives rise to the possibility of the divison of one input PoPS signal into two output signals of different strength (see Applications).
For the implementation of the PoPS amplifier we wanted to use three of those activators (P4 delta was not included because the above paper demonstrated rather weak induction in response to this activator) in combination with the six above promoters.
Implementation
Construction of activator and promoter biobricks
Sequences
The sequence of the promoters was obtained from the above paper and two further papers:
Bacteriophage P2 Late Promoters II Comparison of the Four Late Promoter Sequences; Gail E. Christie1 and Richard Calendar, J. Mol. Biol. (1985) 181, 373-382
and
Mutational Analysis of a Bacteriophage P4 Late Promoter; GIL B. VAN BOKKELEN, EMILY C. DALE, CONRAD HALLING,AND RICHARD CALENDAR; JOURNAL OF BACTERIOLOGY, Jan. 1991, p. 37-45
The promoter sequences used are:
PF: ttgcccgccttttctttaccggtggttgtgctgtcgattagccaaccgggacaaatagcctgacatctccggcgcaactgaaaataccact
PO: cgcgccccgcgattcgctaaggtgctgttgtgtcagtgataagccatccgggactgatggcggaggatgcgcatcgtcgggaaactgatgcc
PP: atgcgcatcctccgccatcagtcccggatggcttatcactgacacaacagcaccttagcgaatcgcggggcgcgactcagtagccttgccgt
PV: ctgaccattacgctctccttgaatgttgtctggtagttctacaaatgaatccagatagcataacttttatatattgtgcaatctcacatgc
Psid: tggcttgccggtctgaggatgagtctcctgtgtcagggctggcacatctgcaatgcgtcgtgttgttgtccggtgtacgtcacaattttctta
PLL: gtggagatgaacaaaaaacacacagccattgtaagacagcctgaacaaatcccccctgttgcgtctgctgaaaatattcacaaaataaagcg
In order to assemble those into biobricks two overlapping oligonucleotides with biobrick prefix(forward)- or suffix(reverse) tails were used. In a PCR process those were annealed and elongated.
As the activator sequences were considerably longer than the promoters shown above, this elongation method could not be used to biobrick them. Instead plasmid material, which was kindly provided by Prof. Richard Calendar, University of Berkeley, California, was used as a template for a PCR to create activator sequences with biobrick ends.
Mutation of the phiR73 delta activator
The original sequence of the phiR73 delta activator contained a PstI restriction site. In order to be used in standard assembly processes it had to be removed. This was successfully achieved by introducing a silent point mutation, using the 2-step megaprimer PCR method.
Resulting biobricks
The biobricks generated are:
Activators
I746350: RBS - P2 ogr activator
I746351: RBS - PSP3 pag activator
I746352: RBS - phiR73 delta activator (with the PstI site successfully removed)
Promoters
I746360: PF promoter
I746361: PO promoter
I746362: PP promoter
I746364: Psid promoter
I746365: PLL promoter
Three activators and five the promoters were successfully biobricked. These biobricks were then used to assess the functionality and the properties of those parts.
Screening constructs
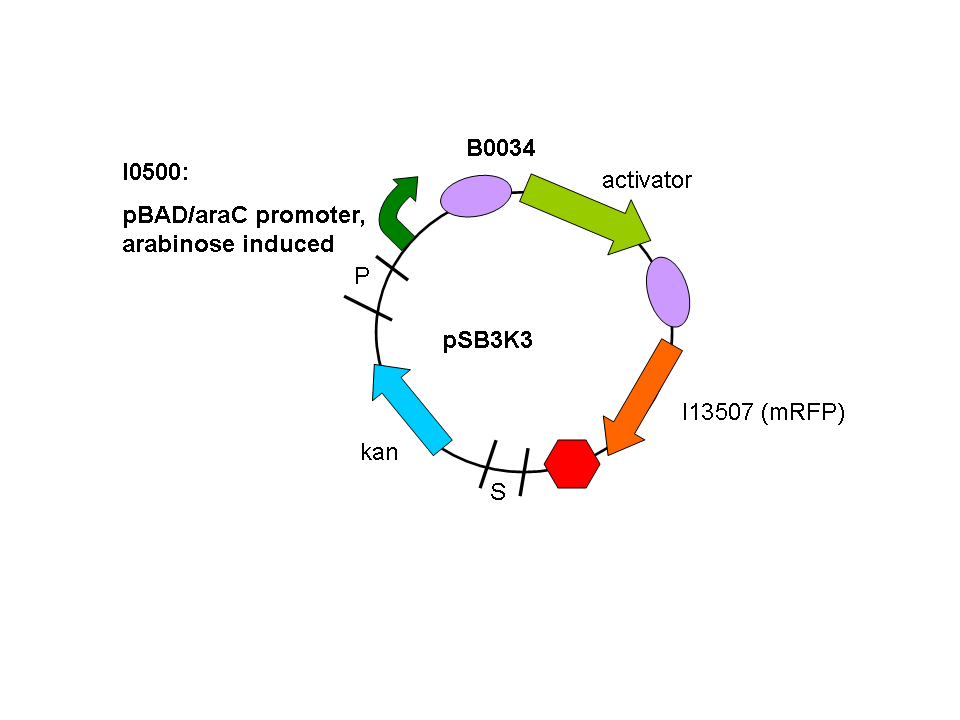
We used a parallel working process to build our constructs, and chose to situate out activator and promoter biobricks on plasmids with different antibiotic resistances (psB1A2 and pSB3K3) to enable us to do some initial screening.
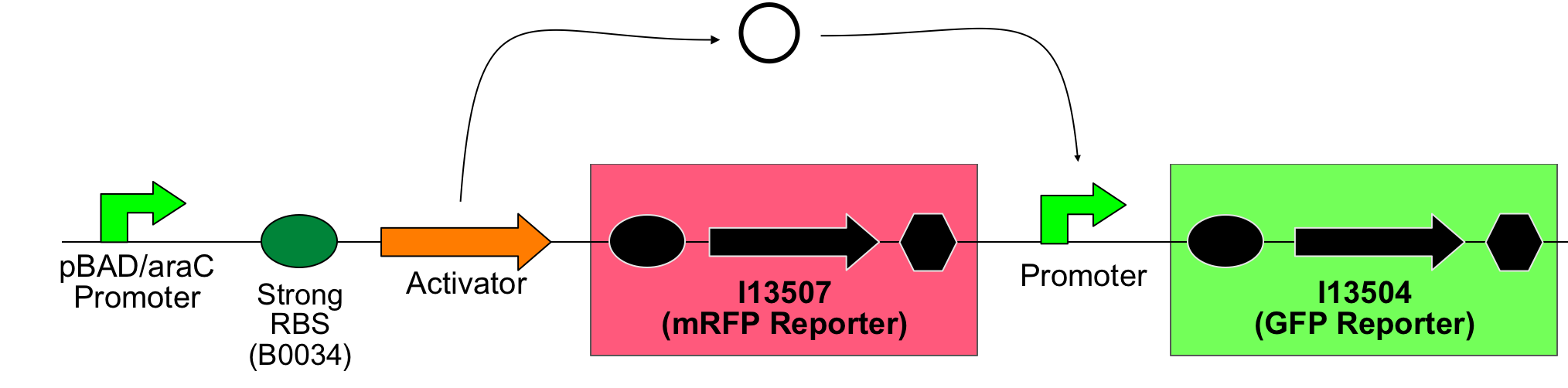
So a first screening to test for induction of the promoters by the activators was done using double transformants. The Ec10G E.cloni strain were first transformed with the following construct which contains the activator and a mRFP reporter under control of the tightly regulated, arabinose induced pBAD/araC promoter:
Biobrick numbers: I746313 (P2 ogr), I746314 (PSP3 pag) and I746315 (phiR73 delta).
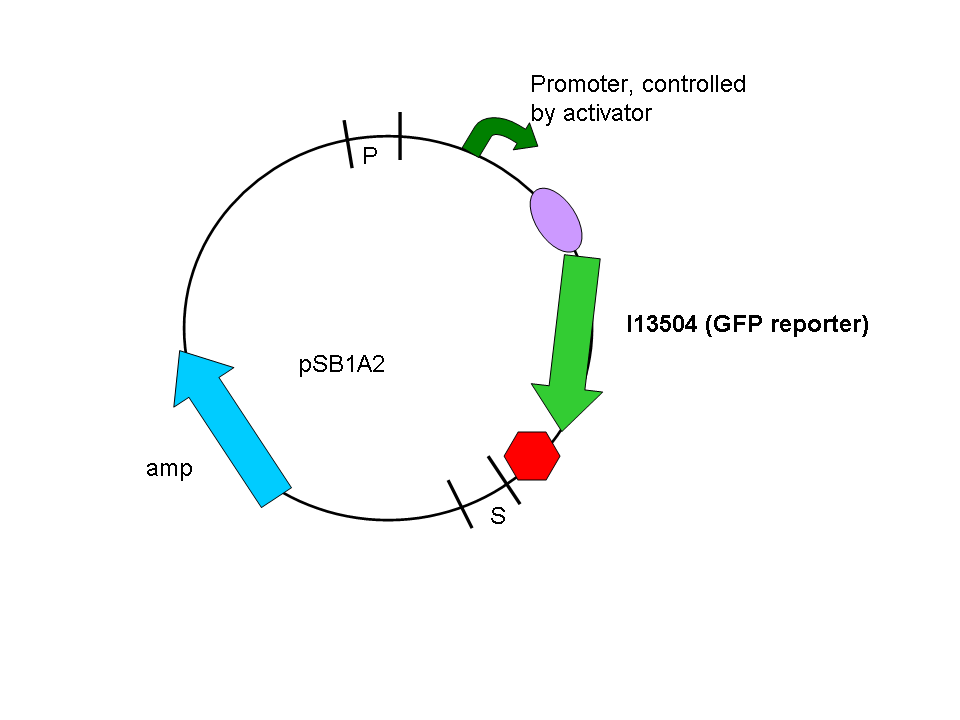
In a second step another plasmid, containing the promoter and a GFP reporter was introduced into the same cells:
Biobrick numbers: I746320(PF), I746321(PO), I746322(PP), I746324(Psid), I746325(PLL)
The cells containing both constructs were found to show arabinose dependent mRFP AND GFP expression, in all possible combinations of activator and promoter (3 activators by 5 promoters = 15 combinations). No fluorescence was observed in non - induced cultures. This indicated that the activators were produced and successfully induced transcription from all the promoters.
As the two plasmids exist in different copy numbers in the cell (pSB1A2 has a high copy number while psB3K3 has a rather low copy number) quantitative results could not be obtained from those constructs. - in order to characterise any possible amplifiers quantitative results were crucial.
Thus, screening constructs with the following basic pattern were built:
All possible combinations of activator and promoter were successfully constructed:
| P2 ogr | PSP3 pag | phiR73 delta | |
| PF promoter | I746370 | I746380 | I746390 |
| PO promoter | I746371 | I746381 | I746391 |
| PP promoter | I746372 | I746382 | I746392 |
| Psid promoter | I746374 | I746384 | I746394 |
| PLL promoter | I746375 | I746385 | I746395 |
Other constructs
In order to assist further constructions using the new promoters and activators the following constructs were built:
- I746340: RBS - P2ogr - B0015
- I746341: RBS - PSP3pag - B0015
- I746342: RBS - phiR73 delta - B0015
Results
- We succesfully produced and tested biobricks of three new activators and five new promoters.
- The activators were shown to induce the promoters to different degrees, thus allowing for the deliberate introduction of crosstalk into systems.
- We obtained quantitative results from experiments on a 96 well plate reader machine. Overnight cultures were diluted, grown into log phase and induced with different concentrations of arabinose.
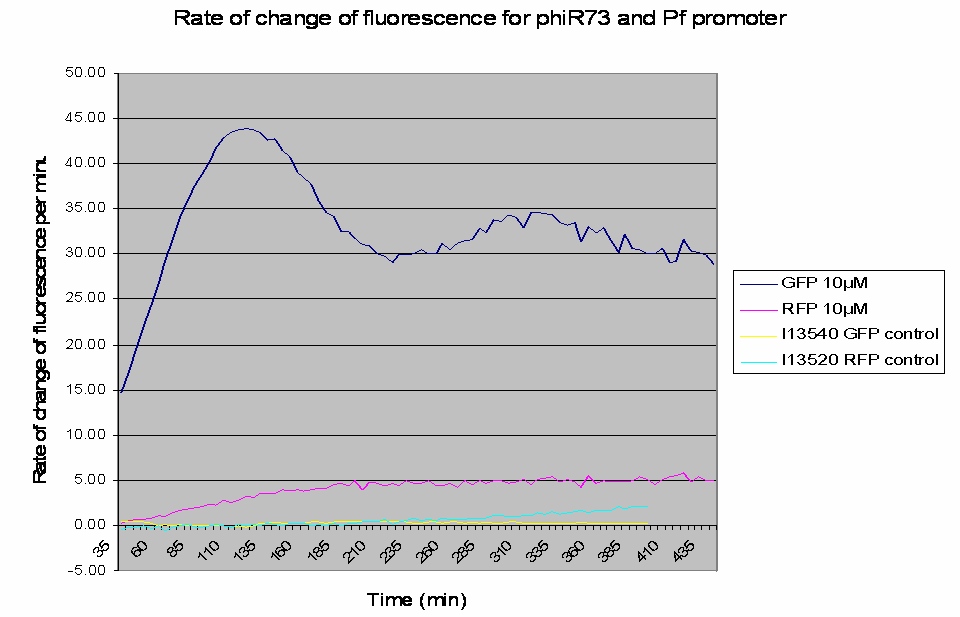
The raw data obtained was processed to analyse the rate of change of the fluorescence levels (this is an adequate representation of PoPS). Example data for one construct (I746390) is shown below: The positive controls used were I13540 (pBAD - GFP) and I13520 (pBAD - mRFP)
Graph shows the rate of change of fluorescence for an amplifier construct containing phiR73 (activator) and Pf (promoter) when induced with 10µM L-Arabinose. Controls are I13520 (Pbad with mRFP) and I13540 (Pbad with GFP). It can be seen that the 'Rate of change of fluorescence' for the amplifier promoter levels at a higher value than the system promoter (ie Pbad shown by the RFP), hence amplification has occurred as there is a higher rate of transcription and thus an increased level of PoPS.
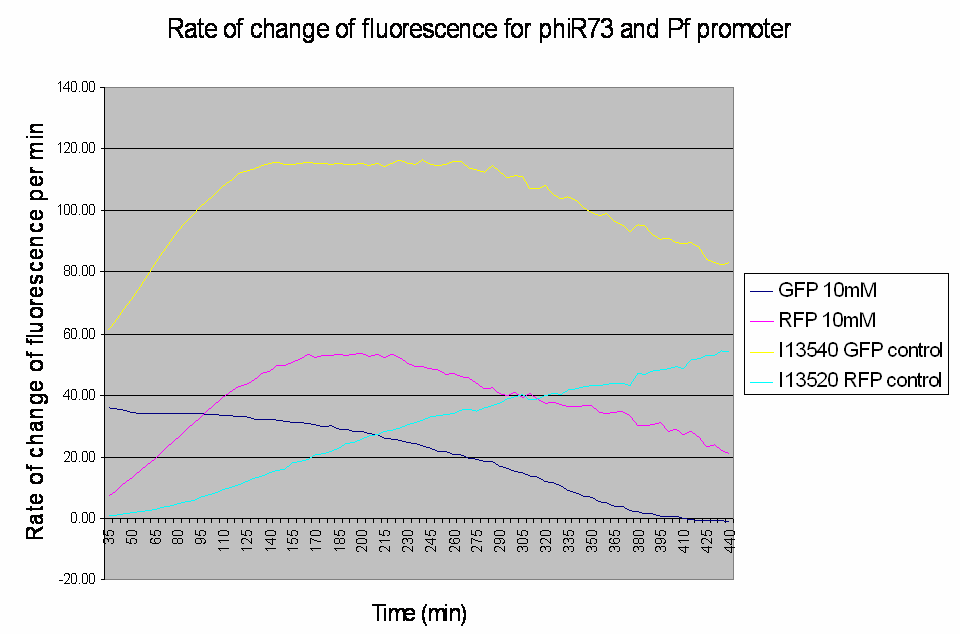
Graph shows the rate of change of fluorescence for an amplifier construct containing phiR73 (activator) and Pf (promoter) when induced with 10mM L-Arabinose. Controls are I13520 (Pbad with mRFP) and I13540 (Pbad with GFP). It can be seen that the 'Rate of change of fluorescence' for the promoter does not level for the amplifier construct, instead it decreases as the rate of change of the activator increases.
The above data illustrates that:
- The promoters are induced by the activators.
- At lower levels of L-Arabinose induction there is amplification by the constructs.
- At high induction levels (i.e. at high rates of transcription from the pBAD promoter) there is no amplification - the observed GFP signal from the construct is much weaker than the control levels from the pBAD promoter.
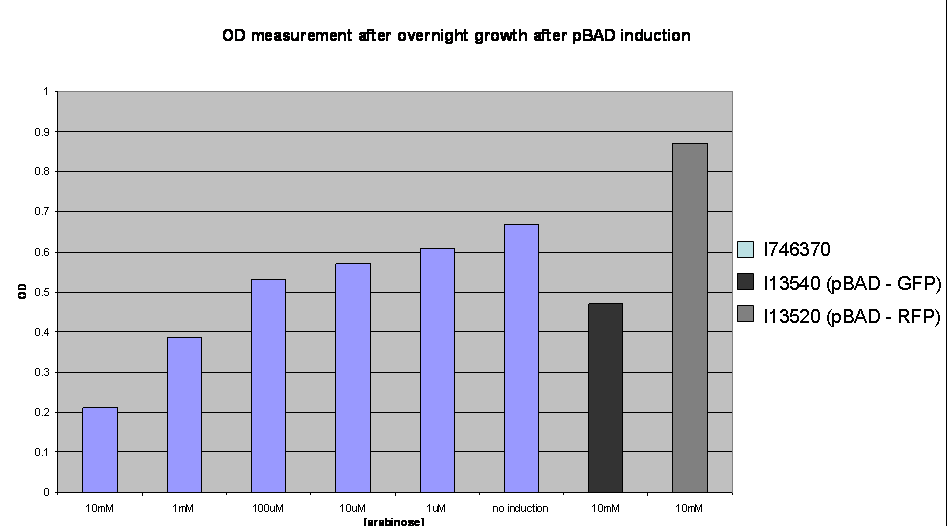
To investigate the nature of this problem OD readings were taken after overnight growth of the cultures after induction:
OD values after overnight incubation: Cultures were induced with arabinose (1: 10mM; 2: 1mM; 3: 100uM; 4: 10uM; 5: 1uM; 6: no induction; 7: I746320, 8: I746340 both induced with 10mM arabinose) and grown overnight before the reading was taken.
This clearly shows a reduced growth of the highly induced cultures. This is likely to account for the low level of induction observed and may be due to high levels of the activator molecules being poisonous to the cell, however, we have not yet investigated this.
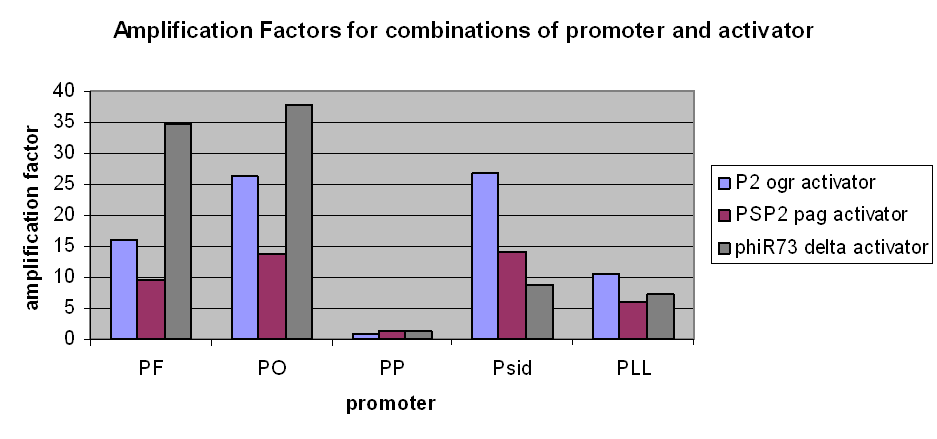
The first graph clearly demonstrates an amplification at LOW levels of arabinose induction. Amplification factors were calculated after 165 minutes of the plate reader experiment after induction with 10-6M arabinose and range from 0.2 to 40:
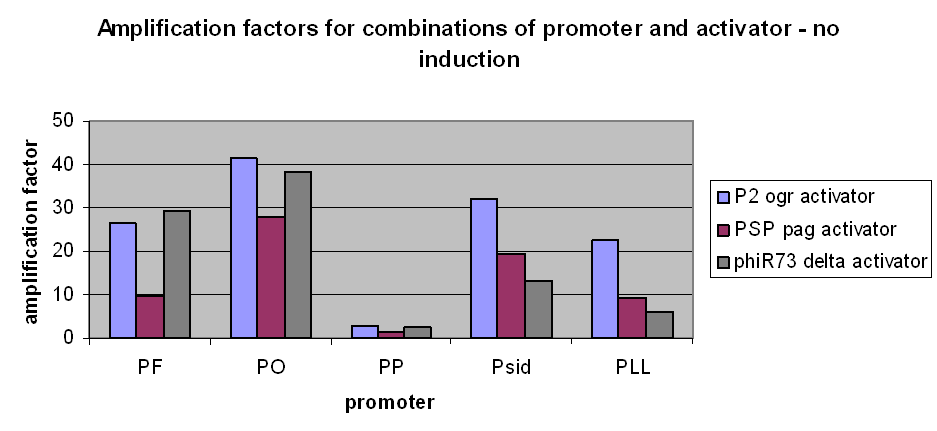
However, we have observed that amplification still occurs at very low levels and very low basal transcriptional activity of the pBAD promoter was also amplified. The amplification factors for this were similar or larger when the pBAD promoter was not induced at all:
Conclusions
- The promoters are induced by the activators.
- At low levels of activator production amplification occurs.
- At higher levels of activator production the observable density of the cells decreases and hence promoter activity decreases.
- Each activator acts on each of the promoters, giving fifteen possible constructs.
- Amplification levels vary between the constructs, and have been calculated.
Applications
The system of transcriptional activators and promoters that was biobricked and characterised has many possible applications in future projects:
- Standard Amplifier:
We have constructed well characterised amplifiers which take the standard PoPS input giving standard PoPS output without interference with other cellular activites. Whilst currently there is an issue with toxicity at higher levels of activator production, it is hoped that this can be solved to give non-limited amplifiers.
- Generating artificial signalling networks:
This application is potentially a very powerful one: A system of intracellular peptide signals combined with promoters with different specificities will be a very useful tool when building complex signalling pathways in a cell: Every natural signalling network relies on regulated crosstalk of its components: Signal division (divergence) and fusion (convergence) are very important concepts that could be realised using a class of intracellular peptide transcriptional activators as shown here.
A simple example would be a signal divider that takes a single PoPS input, dividing it into several PoPS outputs at different levels (as the promoters are induced to different degrees by the same activator).
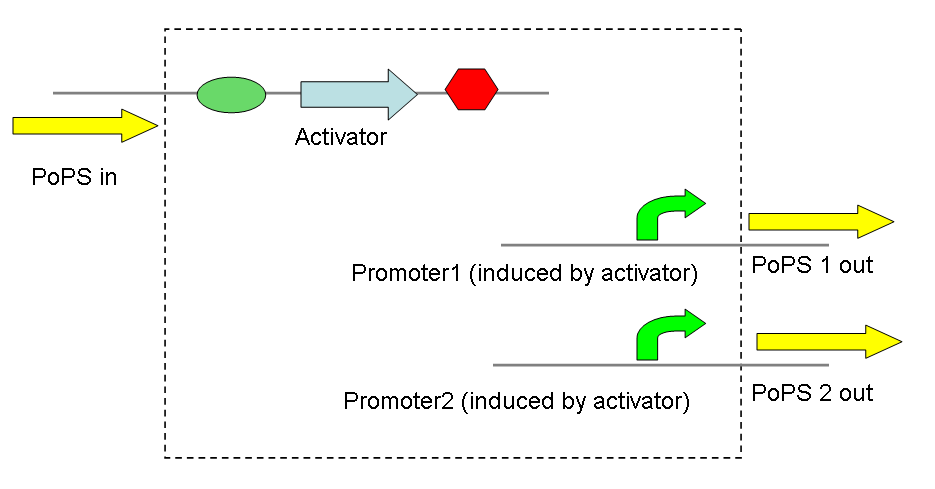
A very similar application is a simple convergence of two PoPS signals, producing 2 activators that act on the same promoter - again to varying degrees. Taken further this could be used to develop a network of transcriptional activators and promoters, thereby allowing complex signal processing.
In the light of our whole project, this network could be used to process the different inputs from several agr-type intercellular signalling systems within each of the cells of the bacterial network.
Future work
A huge amount of data has been gathered to support our conclusions and allow good characterisation of the parts, further data would increase this support, increasing the reliability of the parts.
No work has currently been done on the effects of having two different activators being transcribed in one cell. This could yield some interesting results, especially if the activator binding sites occupy the same space as the resulting promoter activity would depend upon the relative affinities of the two activator molecules for the promoter. This would make a very interesting extension to our work, which unfortunately we were not able to complete due to time constraints.
Current issues
- The reduced growth in highly induced cultures described above is clearly an effect of the transcriptional activators. Our results show a large drop off in observed density as the activator concentration increases. The effect is much more severe than the (slightly) reduced growth in the control cultures (fully induced I13540 and I13520).
- Amplification is only observed at low levels. This clearly relates to the reduced growth of the more highly induced cultures described above - a solution to the growth problems might result in an amplification at higher levels of pBAD transcriptional activity.
- As shown above, even the very low basal level of transcription from the pBAD promoter is amplified up to 40-fold. (Thus demonstrating the sensitivity of the amplification system).
Taken together, those issues indicate that the transcriptional activators are very stable within the cell: Even a low rate of transcription is sufficient to accumulate enough activator to achieve amplified rates of transcription. This stability then means that at high levels a large degree of accumulation of activator must be happening, thus poisoning the cells. So a reasonable first step in addressing the problems described above would be to reduce the halflife of the activator and its mRNA by using appropriate degradation tags - a reasonable balance between degradation and accumulation might result in amplification at a desired level of pBAD transcriptional activity.