Valencia/Welcome to the world of Synthetic Biology and the iGEM competition
From 2007.igem.org
This page is devoted to give our point of view about synthetic biology and the iGEM competition, mainly for those who had never heard about it.
Contents |
What is synthetic biology?
The first steps in Synthetic Biology were done by reaserchers of MIT (motivated by results of many other reaserchers), and grew up as a new (genial) way of seeing genetic engineering.
Genetic engineering is devoted to the manipulation of all molecules and processes involved in the informational metabolism of cells (please visit the informational metabolism site if you know nothing about this, want to refresh your mind, or simply want to know our point of view of the processes involved in). This has been an exhaustively studied area in the last years, and have supposed a revolution in our way of seeing organisms.
The idea of seeing the DNA of organisms as a genetic network where the product of one gene (its expressed protein) can interact with other genes in the network to inhibit or activate the expression of their product is not new (this is just regulation of gene expression but between genes of the same "network"). This is just the beginning point of synthetic biology.
The question is: can we use this way of seeing genetic networks to make biological systems with specified functions? The positive answer to this question relies on the actual capacity of genetic engineering to construct any imaginable sequence of DNA.
Basically, if we can theorically design a genetic network with an specific function (i.e. give the DNA sequence of each gene in the network) we can construct it on laboratory. Then, we can insert this DNA sequence in the cells of an organism and we'll expect from it a "controlled behaviour".
This is the goal not only of synthetic biology but of all areas surrounding genetic engineering. The real power of synthetic biology to achieve this goal relies on the way it approaches the problem.
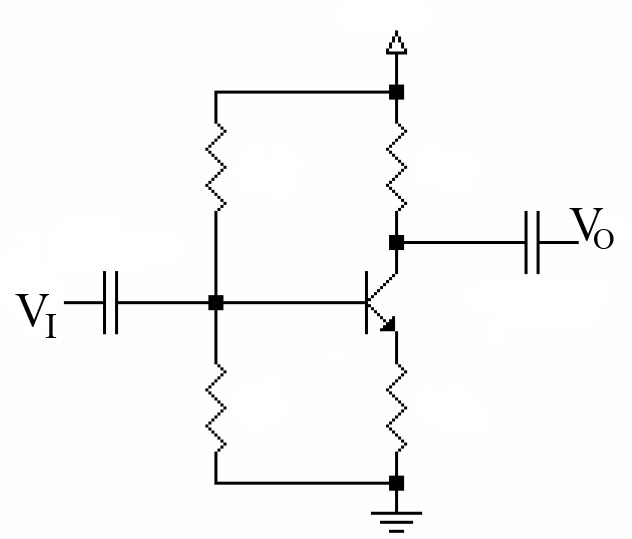
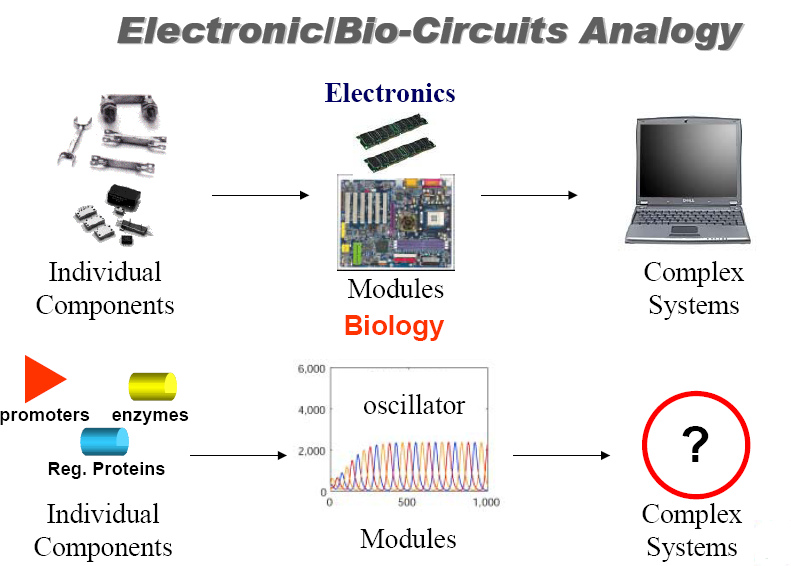
Synthetic biology methods are inspired by the electrical engineering methods. Electronic circuits are composed by little components (diodes, elements, capacitors, etc...) that together can form a device with specific functions (a computer, a DVD, etc...). What is important is that, in order to design this devices, an engineer does not need to know all the complicated electromagnetic processes underlying into those little components, but only how this components respond to an input signal to give an output signal of the same physical characteristics (in this case voltage is the I/O signal). Another important thing is that all this electric components are standarized, so an engineer can use the devices and components designed by other engineers to make his own device.
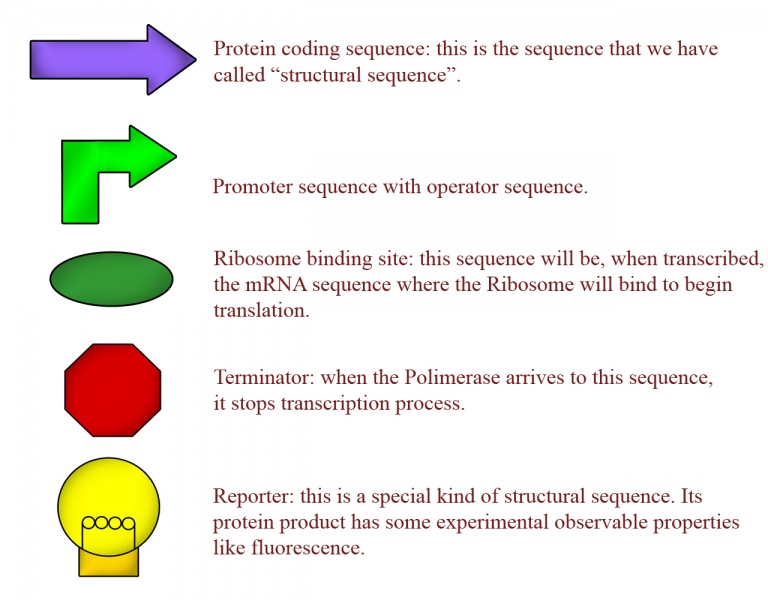
Synthetic biology takes this ideas and adapts them to the designing of new genetic circuits with pre-determined goals. This seems easy because we know many sequences in DNA with specific functions: promoter sequences are recognized by polymerase to begin transcription/replication, terminator sequences can stop transcription, structural sequences are transcribed into mRNA (and then are the precursors of proteins), at the operator sequences many proteins can bind to regulate transcription, and so on... All this DNA sequences with specified function inside the cell are called BioBricks (in the MIT's language), and the graphic symbol for some BioBricks are represented on fig.2.
However, the situation is not as easy as it seems; on electronic circuits the I/O signal is Voltage, and is it for all I/O signals in all electronic components of the circuit. The I/O signal of biologycal components (promoters, terminators, etc...) cannot be the same for all.
This can be understood with a simple example (figs.3a and 3b): we have a gene able to express a protein; in addition, another protein can add to the operator sequence of the gene and act as a repressor.
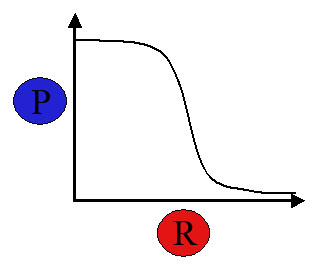
Then this system can be seen as an inverter: when the input signal is high (high repressor concentration), the output signal is low (low expression protein product of gene), as we comment on fig.3c. But here is the main problem: the input signal (repressor protein) and the output signal (the gene product protein) are not the same physical signal (they are not the same protein).
Then, if we want biologycal components to be as interchangeable and universal as electronic components, we have to find a common physical I/O signal for all of these BioBricks. This is one of the central ideas of Sinthetic Biology as understood by MIT's researchers: any sequence of the genetic code is read by polimerase, so polimerase has to be the universal I/O signal. For this purpose they have invented PoPS (Polimeras Per Second), a unit that has to be understood as the number mRNA transcripted per second from the structural sequence of a gene.
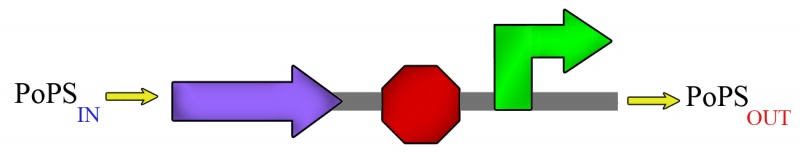
This is clearly shown in the example of fig.4a. The module (we call module to a BioBricks ensemble) starts with a structural sequence; then, if by some mechanism polimerase begins the lecture of the module, the protein corresponding to this structural sequence will be expressed. Now, we finish the module with a promoter/operator sequence regulated by this protein. We can see easily how this module is an inverter with the same physical I/O signal; supose that the polimerase input signal is high, then the protein will be expressed at high amounts, and will bind to the operator sequence at the end of the module to prevent the binding of polimerase. Then, polimerase output signal will be low, because the repression action of the protein. If the input polimerase signal is low, the protein won't be expressed, so polimerase will bind to the pomoter, and the output polimerase signal will be high. This is what we show in fig.4b; please note that both input and output signals are polimerase, i.e. the same physical signal.
This is the central idea of Synthetic Biology. Now the idea is to standarize BioBricks, and for this purpose MIT's researchers have developed the [http://partsregistry.org registry of standard biological parts] site, where all synthetic biology resarchers can add his own designed/created BioBricks and BioBricks ensambles, explaining its function and specifities.
Systems Biology versus Synthetic Biology
Systems Biology is a discipline aiming to understand biology as a system. There we would consider a biological system in Nature and try to modelize it. Using mathematical models and simulation methods can see the temporal reponse system and predict its behavior. Synthetic Biology is an approach which takes the Systems Biology knowledge and try to build new artifial circuits with applications in fields as medecine, energy production... Synthetic Biology is a new discipline born in this millennium. This is a different point of view respect systems biology: this is an engineering approach. We understand biology as technology. We take DNA sequence, called parts by the synthetic biology group at MIT which has recently created the Standard Registry of Parts. They consist of short DNA fragments with an identity, that is, promoters, RBS, coding parts, terminators, etc. With a given function and behaviour and we try to connect them to design a new device or a system with a complex function. We can characterize parts experimentally and simulate this behaviour theoretically. Then we can compare them to construct our genetic system. Unfortunately, the real situation is not as easy as it seems, because of the cellular context in what our biological circuit work is embedded. In electronic circuits, however, each component is connected without thinking in compatibility and isolated from the rest. Nevertheless, in genetic circuits the situation is very different. Inside the cell there are many other molecules that can interact with the genetic sequence that we insert. Furthermore, if one protein is synthetized in some BioBrick of our circuit, it is difused to the cellular medium, being able to interact with anyother BioBrick. Then, in genetic circuits, components are not isolated. For this reason we have to work with BioBricks that does not interact with the cellular medium.
On the other hand, at time to go to the laboratory, we will need a procedure to build our biological system, namely, a procedure to manipulate DNA. We will use standard procedures people use respect manipulation, but we will work with the standard assembling of BioBricks (BioBricks is the name to design parts). For instance, we will need to put in the same plasmid a promoter and next a RBS. For that we will use the following restriction enzymes: EcoRI (E), XbaI (X), SpeI (S) and PstI (P). In addition, XbaI and SpeI have the same sticky ends, so when they assemble make a solid union and a solid sequence. This approach to genetical engineering has two main advantages. The first, we can design with a standard procedure (easier than classical biology approach) genetic sequences whose function inside cells is determined by the behaviour of the BioBricks. The second, we can modelize mathematically the behaviour of the genetic circuit before going to the laboratory. This saves a lot of time and money and add some predictionabilities that had been traditionally far from biology methods.
Finally, when we design a device we have to be conscient that this device can be assembled with other, which can be design by others. So it must some standarization in inputs/outputs. In fact, input should have the same nature that output. But if we consider an inverter, which takes an input signal and produces the opposite output signal (promoter and a coding part), with a repressor and a protein, quickly we see that repressor is not the protein, so the nature is not the same. Nevertheless, we can solve this problem by reorganizing the parts: first coding part, next promoter. Like that, protein is an internal characteristic of the device and the input and the output are the same thing: polymerase (its units are polymerases per second (PoPS)).
Advantages, Applications and Problems.
This approach to genetical engineering has two main advantages:
- We can design in an easy way genetic sequences whose function inside cells is determined by the individual behaviour of the BioBricks is made of.
- As the complexity is reduced, we can modelize mathematically the behaviour of the genetic circuit before going to the laboratory. This will save a lot of time and money, and will add some prediction abilities that has been traditionally far from biology methods.
Given this advantages one have the intuition that many new and amazing applications could be posible. Between this applications there is one specially interesting: we can use this genetic circuits to implement digital computation (we have seen for example a digital inverter in the last section, but it is not so complicated to think about how to implement any logical gate).
Unfortunately, the real situation is not as easy as it seems, because of the cellular context in what our biological circuit is embedded. On electronic circuits, each component is connected to the components we want to, and isolate from the rest and from the external medium (the silico plate).
On genetic circuits the situation is very different. On the cell, there are many other molecules, that can interact with the genetic sequence that we insert. Furthermore, if one protein is sinthetized in some BioBrick of our circuit, it is difused to the cellular medium, being able to interact with any other BioBrick.
Then on genetic circuits, components are not isolated from them nor from the medium. For this reason we have to work with BioBricks that interacts weekly with the cellular medium and within them.
The iGEM competition
iGEM is the acronym for 'international Genetically Engineered Machine competition
iGEM has a double intention: stimulate the research on synthetic biology and form future investigators on this new fresh area.
This was the spirit of the contest when created in 2004 by researchers of MIT. For this purpose, the teams participating on iGEM has to develope a project based on:
- Design a genetic circuit with an interesting function.
- Modelize it mathematically to check it before going to the laboratory.
- Finally, the team has to implement experimentally the designed circuit and check its behaviour.
Previous years results
In this section we are going to talk about some devices or systems taken from literature. We will talk about inverters, switches, oscillators, etc. We are going to use logic gates, assuming we know what is that (for more information about that go to the literature).
One remarcable thing is that in electronics, using only NAND gates or only NOR gates we can achieve the behavior of all kind of logic gates. Then, whatever digital electronic circuit can be designed by these logic gates. In biology, we can not say that a priori because the behavior is not perfect, so we introduce this concept of logic gate only to do a comparison and to do a classification.
In synthetic biology we use promoters as parts and we can change it for another to achieve a better (or simply different) behavior. But to use them we need to know how it acts when there are proteins that are be able to bind with the promoter operators, such as repressors or activators. Then, according to the type of protein the system will change. Typically, promoter region is modeled as a logic gate. Then, if we are a promoter upstream a coding region and two repressors that functions, for example, as a NOR gate, namely, if one repressor concentration is high, protein concentration is low; only when there is no repressor there is protein.
A biochemical inverter is a DNA sequence with a promoter (site where RNA Polymerase is fixed) and a protein coding region (site that contains a sequence to transcribe to mRNA in order to product protein) and a repressor (or inductor). Promoter may be repressed or induced by a protein, namely, if a protein acts repressing the promoter active part (operator), RNA Polymerase can not be fixed and then protein is not formed. On the other hand, if protein acts inducing the promoter operator, RNA Polymerase can fix to DNA sequence. If there is not protein to induce, protein is not formed.
To continue with logical devices, we consider now a RS Latch implemented with NAND logic gates (or RS NAND Latch). We can also implement an analog circuit using NOR logic gates (RS NOR Latch) with the same disposition, but changing the logic gates. This is a circuit commonly used in electronics.
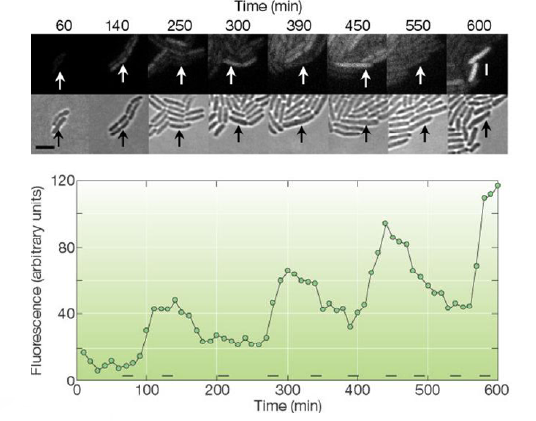
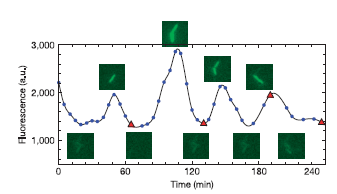
The oscillator, also called repressilator, is a device formed with three NOT logic gates in series. If we calculate the concentrations in midpoints, we can see that concentrations are periodic signals and there is a phase between successive stages. In fact, this system is now implemented in data bank of MIT parts. The reference is I5610.
Using the same approach than in repressilator to obtain an oscillatory circuit, but in this case to metabolic pathways, scientists from University of California - Los Angeles constructed the Metabolator. This circuit has an oscillatory dynamics profiting the path between the acetyl-CoA and the AcP - acetate equilibrium.
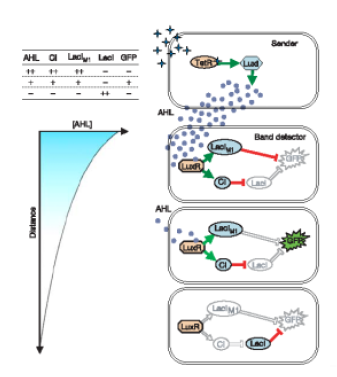
We can see an implementation of this system using AHL as inducer. Weiss' group in Princeton has developed a genetic circuit in order to detect an intermediate concentration level of a transcription factor (in that case AHL). We can obtain that behavior using two branches, one positive, and the other one, negative.