Paris/July 20
From 2007.igem.org
< Paris(Difference between revisions)
(→E.Coli pKS::DGAT) |
Nicolas C. (Talk | contribs) |
||
| (2 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Paris/July 19|yesterday]] -- [[Paris/July 21|tomorrow]] <br> | ||
==PCRs== | ==PCRs== | ||
| Line 60: | Line 61: | ||
* Interpretation | * Interpretation | ||
| + | |||
| + | We can observe lipid inclusions within colonies with DGAT. Further investigations need single cell visualisation. | ||
Latest revision as of 17:49, 7 October 2007
Contents |
PCRs
| PCR : Lox71-KmR-Lox66 | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 1084 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 10 Lox71-KmR-F | 2.5µL | ? | ||
| 3m00' | Oligo R 10µM | 11 Lox66-KmR-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | MP3_pSB1AK3-BBa_B0015 0.5µl | |||||
| PCR : Lox71-FtsA-FtsZ | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 2588 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 3 Lox71-FtsA-F | 2.5µL | ? The PCR seemed to have worked but the purification failed | ||
| 3m00' | Oligo R 10µM | 2 FtsZ-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | toothpick in MG1655 glycerol | |||||
Glycerol stock
- BBa_pJ23107 clone 3&4 (S14)
- BBa_I0500 clone 1&2 (S13)
Minipreps on Biobricks
- BBa_pJ23107 clone 3&4
- BBa_I0500 clone 1&2
E.Coli pKS::DGAT
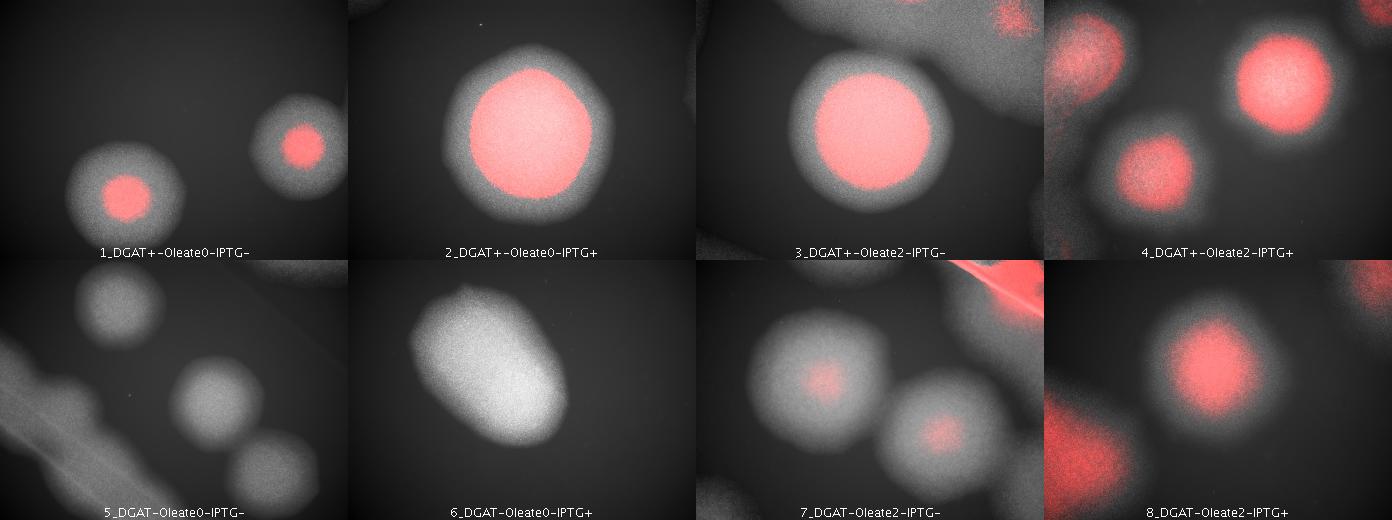
We look under microscopy 36 hours after incubation of E.coli transformed by pKS::DGAT and the control E.coli transformed by part B0015 on different LB medium (See July 18).
- Observations:
We can observe colonies (X10).
- Interpretation
We can observe lipid inclusions within colonies with DGAT. Further investigations need single cell visualisation.