Edinburgh/Yoghurt/Modelling
From 2007.igem.org
| (26 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
---- | ---- | ||
[[Image:Edinburgh Uni Logo.jpg|50 px]] | [[Image:Edinburgh Uni Logo.jpg|50 px]] | ||
| - | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/References|References]] | + | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/Proof of concept|Proof of concept]] | [[Edinburgh/Future| Future Directions]] | [[Edinburgh/Yoghurt/References|References]] |
---- | ---- | ||
| + | ===Vanillin Pathway=== | ||
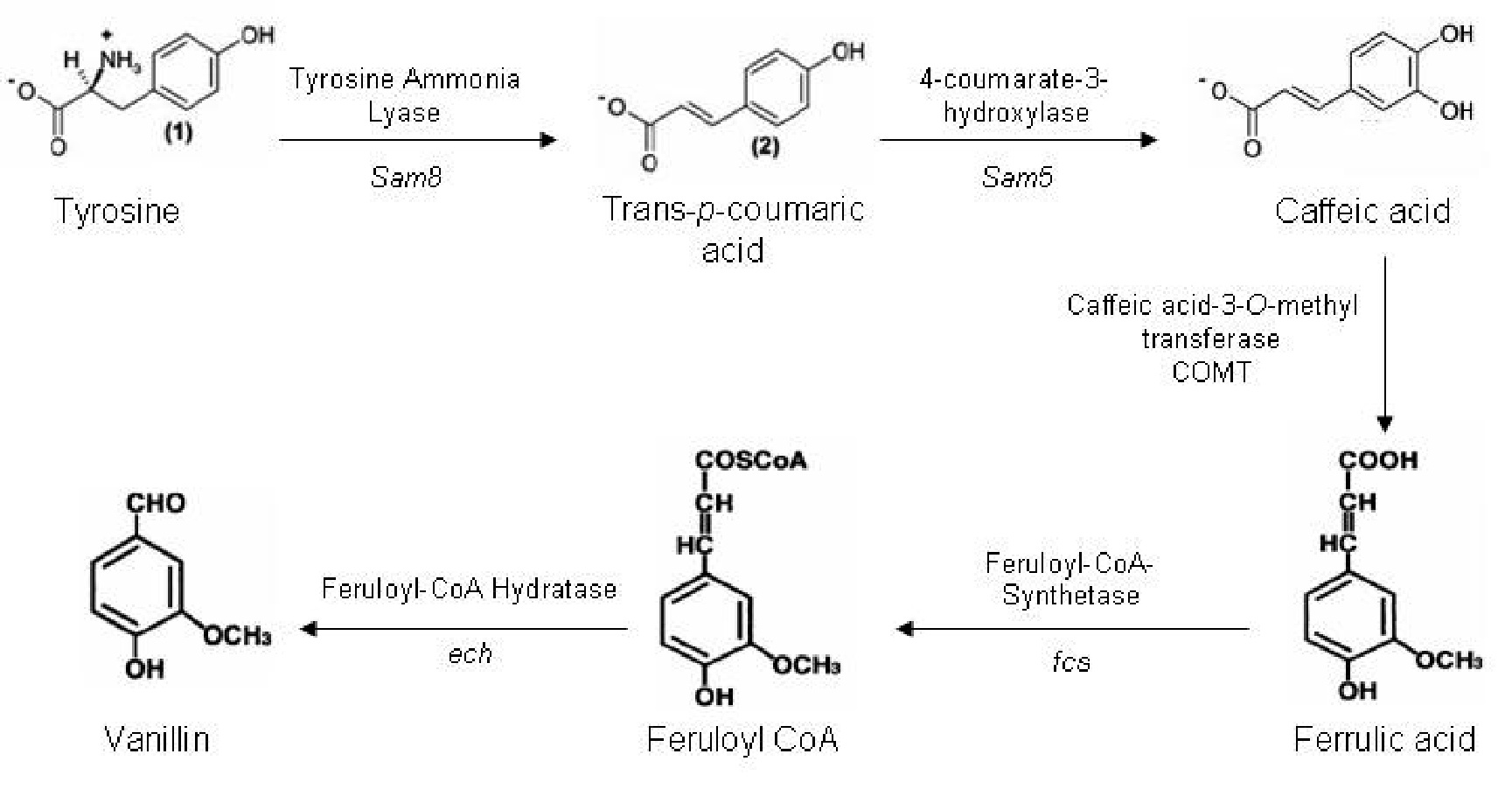
| + | The synthetic pathway to produce vanillin from the amino acid tyrosine is shown below (Fig. 1). There are five genes in this pathway, which are present in three different organisms. | ||
| + | |||
| + | '''Figure 1. The synthesis pathway of vanillin production. (1)''' | ||
| + | |||
| + | [[Image:Fig1_yogurt.jpg|600px]] | ||
| + | |||
| + | The ''Sam5'' and ''Sam8'' genes are from the microorganism ''Saccharothrix espanaensis''. ''Sam8'' encodes a tyrosine ammonia lyase enzyme, and ''Sam5'' a hydroxylase enzyme. The caffeic acid-3-0- methyltransferase (COMT) gene is part of the lignin biosynthesis pathway present in plants. Both ''fcs'' and ''ech'' genes are present in a variety of microorganisms. After much searching we have identified ''ech'' and ''fcs'' genes in ''Pseudomonas fluorescens'', which have a minimal number of forbidden restriction sites. | ||
| + | |||
| + | ===Model of the vanillin pathway=== | ||
| + | So far, we have constructed a basal model of the vanillin pathway, based on several assumptions. The first and most important one is that all of the five enzymes related in the pathway are expressed at sufficient levels. In other words, we did not add the enzyme expression into this initial model. This is different from the situation in the wet lab, but reasonable because the process of vanillin synthesis occurs over a much longer time course than that required for enzyme expression. Thus, the concentrations of enzymes can reach a certain level which is sufficient for vanillin synthesis at the very beginning of the whole pathway. Besides this, we also assume that the concentration of the first substrate, Tyrosine, is fixed at a certain level and there is a consumption system for vanillin. | ||
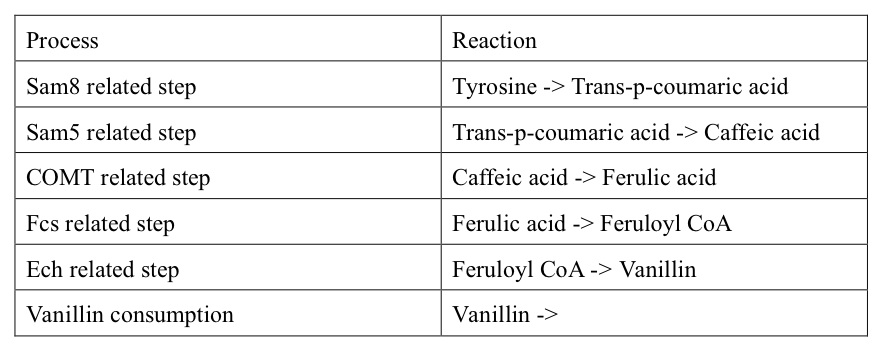
| + | Therefore, the reactions of vanillin pathway in Copasi are listed below: | ||
| + | |||
| + | '''Table1. The reactions in the vanillin pathway''' | ||
| + | |||
| + | [[Image:Tablemodellingyogurt.jpg|600px]]. | ||
| + | |||
| + | |||
| + | ===Results=== | ||
| + | |||
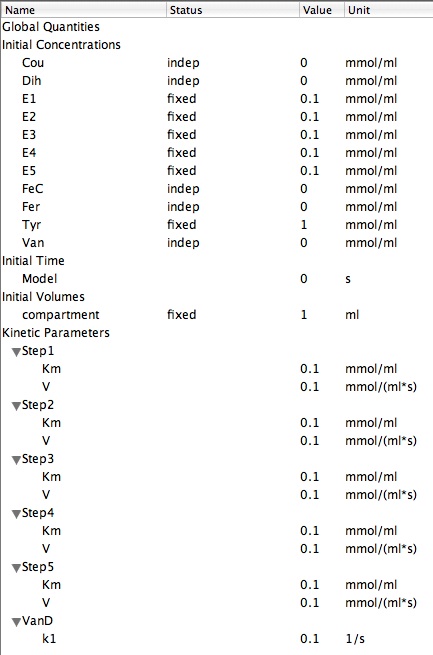
| + | '''The modelling parameters used:''' | ||
| + | |||
| + | [[Image:Model_parameters_yogurt.jpg]] | ||
| + | |||
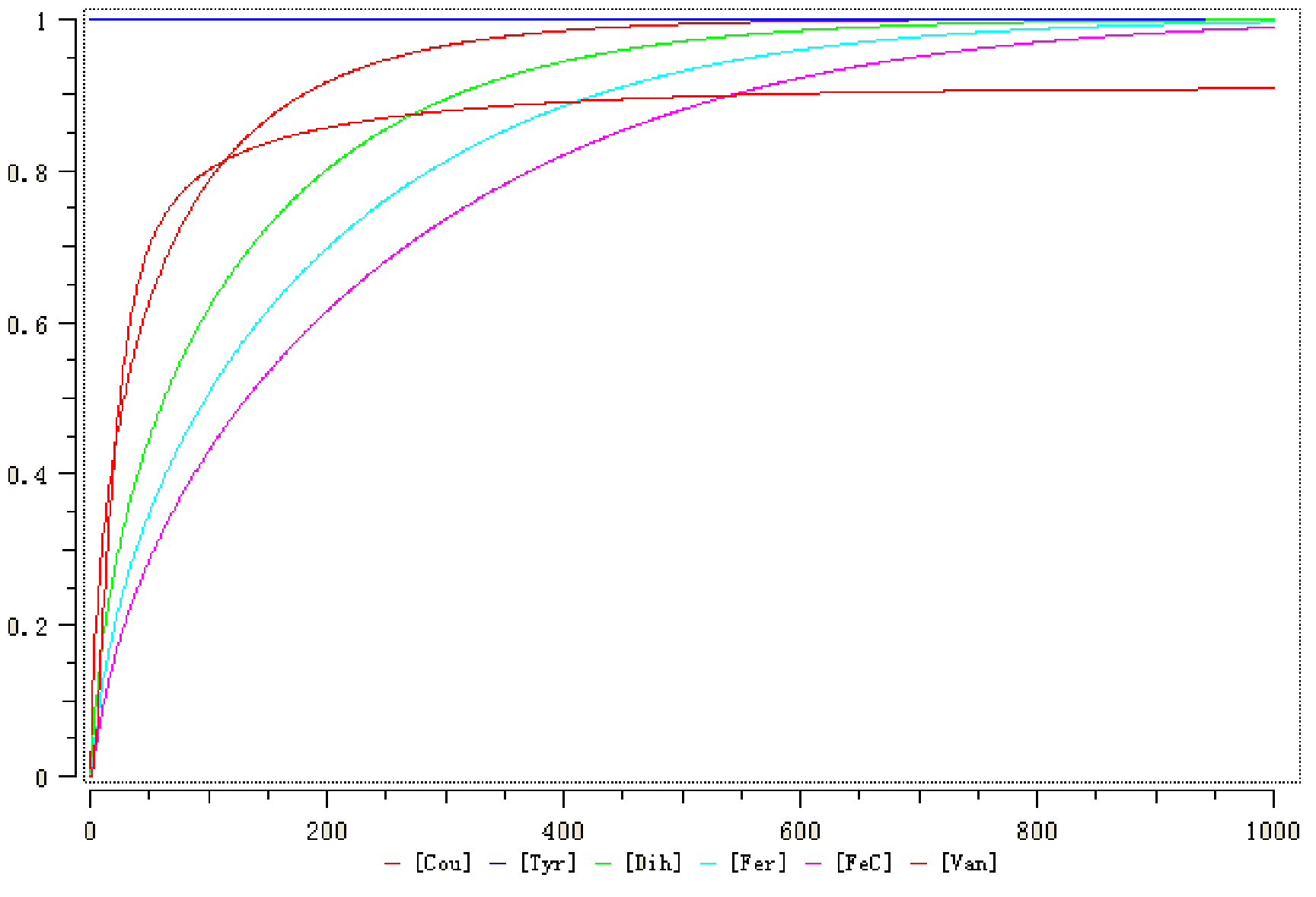
| + | The rates of the first five reactions are Michaelis-Menten equation and Mass equation for the vanillin consumption. The simulation result is shown below (Fig. 2). After several time units, the concentration of valillin (red curve) will reach a steady state (around 0.9 mmol/l). | ||
| + | |||
| + | '''Figure 2. The concentration of substances in vanillin pathway''' | ||
| + | |||
| + | [[Image:Vanillin_modelling.jpg|600px]] | ||
| + | |||
| + | Unit: Concentration: mmol/ml; Time: s. | ||
| + | |||
| + | ===6.5 Future work=== | ||
| + | Since this is only a very simple model, we will add some more realistic structures into it. As mentioned before, the process of enzyme expression is also part of the work in the wet lab. Although it occurs over a shorter time course than vanillin synthesis, it will affect the pathway. Thus, modeling enzyme expression would be our next step (Fig. 3). | ||
| + | |||
| + | ''' Figure3. The vanillin pathway with enzyme expression''' | ||
| + | [[Image:Enzyme_pathway.jpg|600px]] | ||
| + | |||
| + | ===Reference:=== | ||
| + | 1. https://2007.igem.org/Edinburgh. Accessed on August 2007. | ||
---- | ---- | ||
| - | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/References|References]] | + | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/Proof of concept|Proof of concept]] | [[Edinburgh/Future| Future Directions]] | [[Edinburgh/Yoghurt/References|References]] |
---- | ---- | ||
Latest revision as of 20:11, 26 October 2007
 Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | Future Directions | References
Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | Future Directions | References
Contents |
Vanillin Pathway
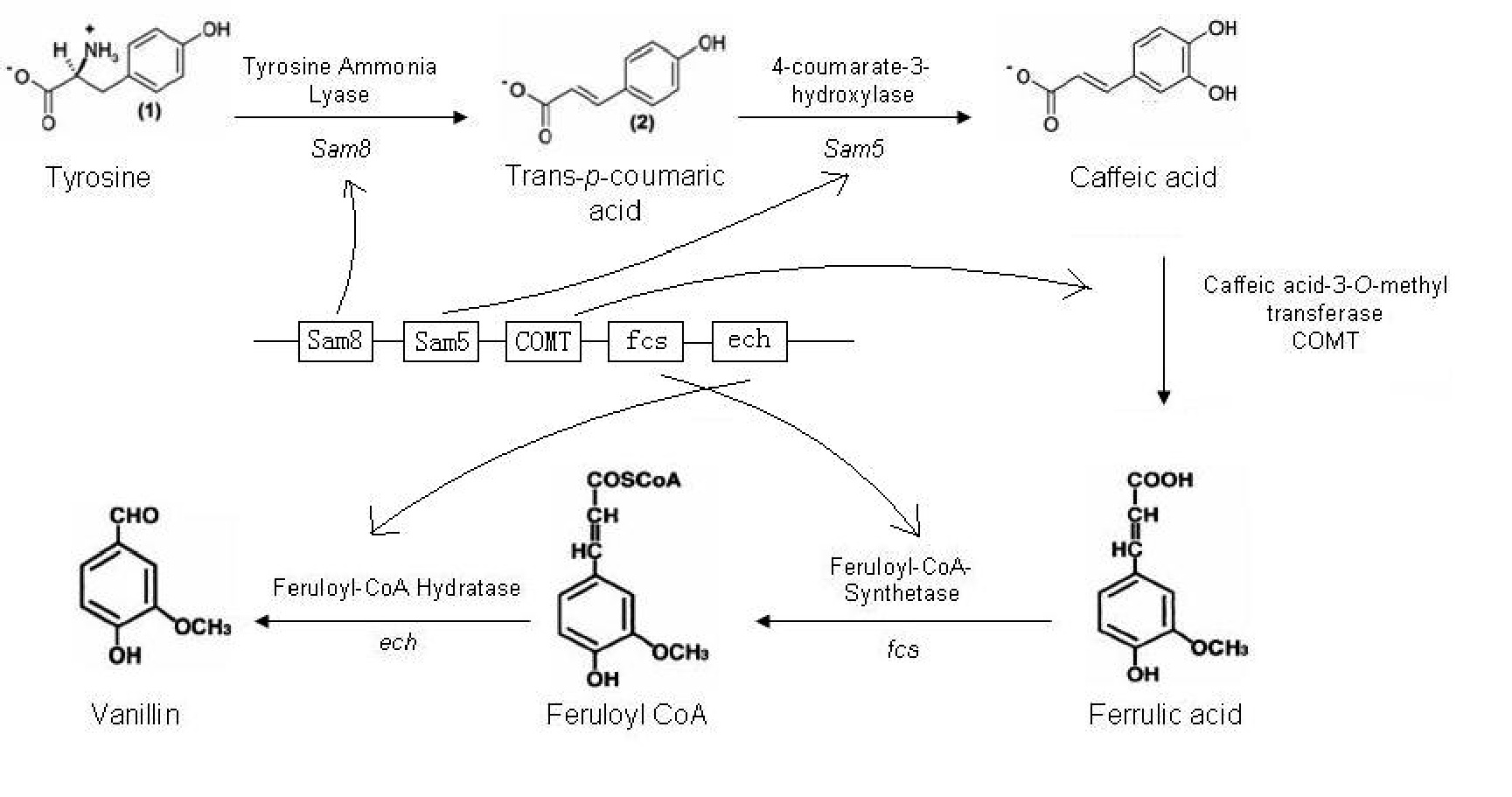
The synthetic pathway to produce vanillin from the amino acid tyrosine is shown below (Fig. 1). There are five genes in this pathway, which are present in three different organisms.
Figure 1. The synthesis pathway of vanillin production. (1)
The Sam5 and Sam8 genes are from the microorganism Saccharothrix espanaensis. Sam8 encodes a tyrosine ammonia lyase enzyme, and Sam5 a hydroxylase enzyme. The caffeic acid-3-0- methyltransferase (COMT) gene is part of the lignin biosynthesis pathway present in plants. Both fcs and ech genes are present in a variety of microorganisms. After much searching we have identified ech and fcs genes in Pseudomonas fluorescens, which have a minimal number of forbidden restriction sites.
Model of the vanillin pathway
So far, we have constructed a basal model of the vanillin pathway, based on several assumptions. The first and most important one is that all of the five enzymes related in the pathway are expressed at sufficient levels. In other words, we did not add the enzyme expression into this initial model. This is different from the situation in the wet lab, but reasonable because the process of vanillin synthesis occurs over a much longer time course than that required for enzyme expression. Thus, the concentrations of enzymes can reach a certain level which is sufficient for vanillin synthesis at the very beginning of the whole pathway. Besides this, we also assume that the concentration of the first substrate, Tyrosine, is fixed at a certain level and there is a consumption system for vanillin. Therefore, the reactions of vanillin pathway in Copasi are listed below:
Table1. The reactions in the vanillin pathway
Results
The modelling parameters used:
The rates of the first five reactions are Michaelis-Menten equation and Mass equation for the vanillin consumption. The simulation result is shown below (Fig. 2). After several time units, the concentration of valillin (red curve) will reach a steady state (around 0.9 mmol/l).
Figure 2. The concentration of substances in vanillin pathway
Unit: Concentration: mmol/ml; Time: s.
6.5 Future work
Since this is only a very simple model, we will add some more realistic structures into it. As mentioned before, the process of enzyme expression is also part of the work in the wet lab. Although it occurs over a shorter time course than vanillin synthesis, it will affect the pathway. Thus, modeling enzyme expression would be our next step (Fig. 3).
Figure3. The vanillin pathway with enzyme expression
Reference:
1. https://2007.igem.org/Edinburgh. Accessed on August 2007.
Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | Future Directions | References