Freiburg07/report light sensor
From 2007.igem.org
| (34 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<h2>Introduction</h2> | <h2>Introduction</h2> | ||
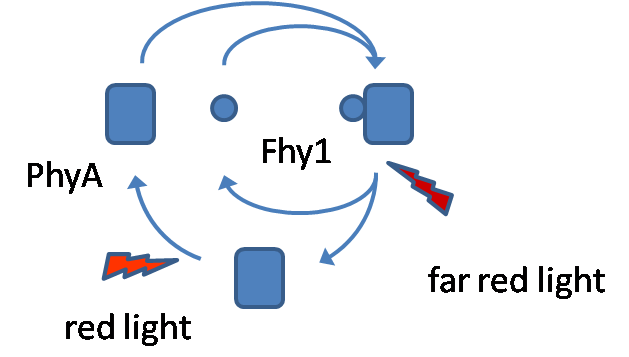
| - | Phytochrome A (PhyA) is a member of the phytochrome family of red/far-red photoreceptors. These photoreceptors are involved in a wide range of developmental responses in plants. Fhy1 is a protein which binds PhyA under light conditions, enable us to use this protein-protein interaction. In plants Fhy1 is | + | Here we want to introduce a possibility for an in vivo light switch. Dependent on the radiation of different wavelengths our system can switch between on and off. One wavelength can switch protein interaction on (red light) and another wavelength can switch it off (far-red light). Therefore we choose a light sensing protein (Phytochrome A) and a protein (Fhy1) binding the light sensor in case of certain light conditions. |
| + | |||
| + | Phytochrome A (PhyA) is a member of the phytochrome family of red/far-red photoreceptors. These photoreceptors are involved in a wide range of developmental responses in plants. It changes between two stable conformations, the Pr (red light absorbing) and Pfr (far-red light absorbing) form. Red light causes a conformational switch into the Pfr form and far-red light switches PhyA into the Pr form (Fhy1 is bound by the Pr form). The conformational change is induced by PCB, the chromophore of PhyA, a tetrapyrrole which performs a rotation around a carbon bound by light radiation. PCB does naturally not exist in E. coli, that is why an extra plasmid with PCB synthesis enzymes is transformed parallel with a plasmid containing PhyA-CFP and another one with Fhy1-YFP. Fhy1 is a protein which binds PhyA under red light conditions, enable us to use this protein-protein interaction. In plants Fhy1 is responsible for nuclear accumulation of PhyA in case of light exposure. Nuclear transport is essential for photo-responses, because PhyA acts as transcription factor. In our case it was not necessary to use the full Fhy1 protein, that is why we restricted to the PhyA binding domain of Fhy1. We do not need the full Fhy1 peptide because nuclear accumulation is not necessary in our case. We choose two different sizes of the Fhy1 fragment, a small version and a big one. Our goal was to show the interaction of both by performing a [http://en.wikipedia.org/wiki/Fluorescence_resonance_energy_transfer FRET], fusing CFP to PhyA and YFP to Fhy1. | ||
<div style="left"> | <div style="left"> | ||
| Line 19: | Line 21: | ||
Fhy1 Insert1 was gained by PCR from pGADT7 using 5´ (5´-TATCGACGCT AGCGATTACA TCTATGGGAC TCAGA-3´) and 3´<br> (5´-AACGATCGGC GCGCCCAGCA TTAGCGTTGA GAAGTAT-3´) primers.<br> | Fhy1 Insert1 was gained by PCR from pGADT7 using 5´ (5´-TATCGACGCT AGCGATTACA TCTATGGGAC TCAGA-3´) and 3´<br> (5´-AACGATCGGC GCGCCCAGCA TTAGCGTTGA GAAGTAT-3´) primers.<br> | ||
Fhy1 Insert2 was gained by PCR from pGADT7 using 5´ (5´-TATCGACGCT AGCAGTAGTA ATGCTGCTAA GTTTGT-3`) and 3´<br> (5´-AACGATCGGC GCGCCCAGCA TTAGCGTTGA GAAGTAT-3´) primers.<br> | Fhy1 Insert2 was gained by PCR from pGADT7 using 5´ (5´-TATCGACGCT AGCAGTAGTA ATGCTGCTAA GTTTGT-3`) and 3´<br> (5´-AACGATCGGC GCGCCCAGCA TTAGCGTTGA GAAGTAT-3´) primers.<br> | ||
| + | |||
| + | PhyA Insert was digested with EcoRI and SpeI, the CFP containing plasmid pAR200-cJunCFP as well. | ||
| + | Fhy1 Insert1 was digested with AscI and NheI, the YFP containing plasmid pAR200-YFP-Dummy (for N-terminal fusion) and pAR200-cFOS-YFP (for C-terminal fusion) as well. All vector plasmids were provided by our laboratory. | ||
| + | |||
| + | Insert and Vector fragments were ligated. The purified DNA was used for a transformation in E. coli. | ||
| + | The E. coli DNA was purified and sequenzed. | ||
| + | |||
| + | [[Freiburg07/Mediums and Plates| Mediums and Plates]]<BR> | ||
| + | [[Freiburg07/DNA_sequencing| DNA sequencing]]<BR> | ||
| + | [[Freiburg07/Ligation| Ligation]]<BR> | ||
| + | [[Freiburg07/Plasmidprep| Plasmid spin column prep]]<BR> | ||
| + | [[Freiburg07/Glycerol_stock| Glycerol stocks]]<BR> | ||
| + | [[Freiburg07/Gene_Protein_info| General Gene-Protein Information]]<BR> | ||
| + | [[Freiburg07/Purification|Purification ]]<BR> | ||
| + | [[Freiburg07/Transformation|Transformation ]]<BR> | ||
| + | [[Freiburg07/Dephosphorylation|Dephosphorylation ]]<BR> | ||
| + | [[Freiburg07/Preparative Digestion|Preparative Digestion ]]<BR> | ||
| + | [[Freiburg07/Analytic Digestion|Analytic Digestion ]]<BR> | ||
| + | [[Freiburg07/Gel Electrophoresis|Gel Electrophoresis ]]<BR> | ||
| + | [[Freiburg07/Polyacrylamide gel electrophoresis|Polyacrylamide gel electrophoresis ]]<BR> | ||
<h2>Results</h2> | <h2>Results</h2> | ||
| + | |||
| + | Plasmids with PhyA and Fhy1 were cloned. Unfortunately our time was running out and we only gained plasmids with mutations yet. | ||
<h2>Discussion</h2> | <h2>Discussion</h2> | ||
| + | |||
| + | The PhyA - Fhy1 light sensing system is a promising approach for regulating protein/enzyme activity by light stimulation. | ||
| + | Of course we need evidence for the correct arrangement, given by the CFP and YFP fusion. Gaining a FRET would help us to be sure about the arrangement for split enzymes. These parts could be fused to PhyA and Fhy1 and get a complementation of the enzyme by light. | ||
| + | In our Fusion Parts-Kit, two different split enzymes are provided (DHFR: BBa_1757005 and BBa_1757006; ß-Lactamase: BBa_1757011 and BBa_1757012). Both could be fused to PhyA and Fhy1, as well as CFP (Cerulean) and YFP (Venus). | ||
| + | |||
| + | <h2> [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Freiburg Parts] </h2> | ||
<h2>References</h2> | <h2>References</h2> | ||
| + | |||
| + | Hiltbrunner A, Tscheuschler A, Viczian A, Kunkel T, Kircher S, Schäfer E. "FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor." Plant Cell Physiol. 2006 Aug;47(8):1023-34. | ||
| + | |||
| + | Hiltbrunner A, Viczian A, Bury E, Tscheuschler A, Kircher S, Toth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E "Nuclear accumulation of the phytochrome A photoreceptor requires FHY1." Curr Biol. 2005 Dec 6;15(23):2125-30. | ||
| + | |||
| + | Gambetta GA, Lagarias JC "Genetic engineering of phytochrome biosynthesis in bacteria." Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10566-71.<br> | ||
| + | |||
| + | Takeharu Nagai, Keiji Ibata, Eun Sun Park, Mie Kubota, Katsuhiko Mikoshiba, Atsushi Miyawaki "A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications." Nature Biotechnology 20, 87 - 90 (01 Jan 2002) Research<br> | ||
| + | |||
| + | Mark A Rizzo, Gerald H Springer, Butch Granada & David W Piston "An improved cyan fluorescent protein variant useful for FRET." Nature Biotechnology 22, 445 - 449 (01 Apr 2004) Research | ||
| + | |||
[https://2007.igem.org/Freiburg BACK] | [https://2007.igem.org/Freiburg BACK] | ||
Latest revision as of 22:06, 26 October 2007
Contents[hide] |
Towards optical signal processing in E. coli
iGEM team Freiburg
Introduction
Here we want to introduce a possibility for an in vivo light switch. Dependent on the radiation of different wavelengths our system can switch between on and off. One wavelength can switch protein interaction on (red light) and another wavelength can switch it off (far-red light). Therefore we choose a light sensing protein (Phytochrome A) and a protein (Fhy1) binding the light sensor in case of certain light conditions.
Phytochrome A (PhyA) is a member of the phytochrome family of red/far-red photoreceptors. These photoreceptors are involved in a wide range of developmental responses in plants. It changes between two stable conformations, the Pr (red light absorbing) and Pfr (far-red light absorbing) form. Red light causes a conformational switch into the Pfr form and far-red light switches PhyA into the Pr form (Fhy1 is bound by the Pr form). The conformational change is induced by PCB, the chromophore of PhyA, a tetrapyrrole which performs a rotation around a carbon bound by light radiation. PCB does naturally not exist in E. coli, that is why an extra plasmid with PCB synthesis enzymes is transformed parallel with a plasmid containing PhyA-CFP and another one with Fhy1-YFP. Fhy1 is a protein which binds PhyA under red light conditions, enable us to use this protein-protein interaction. In plants Fhy1 is responsible for nuclear accumulation of PhyA in case of light exposure. Nuclear transport is essential for photo-responses, because PhyA acts as transcription factor. In our case it was not necessary to use the full Fhy1 protein, that is why we restricted to the PhyA binding domain of Fhy1. We do not need the full Fhy1 peptide because nuclear accumulation is not necessary in our case. We choose two different sizes of the Fhy1 fragment, a small version and a big one. Our goal was to show the interaction of both by performing a [http://en.wikipedia.org/wiki/Fluorescence_resonance_energy_transfer FRET], fusing CFP to PhyA and YFP to Fhy1.
Materials and Methods
Cloning of constructs:
PhyA Insert was gained by PCR from plasmid D153ah-phyA using 5´ (5´-TATCGACGAA TTCGCGGCCG CTTCTAGAAA GAGGAGAAAT
TAACTATGTC AGGCTCTAGG CCGAC-3´) and 3´ (5´- AACGATCACT AGTTCCGCCG TGATGGTGAT GGTGGCCTCC GTTATCGAGT TCCACCTCCT TA-3´) primers.
Fhy1 Insert1 was gained by PCR from pGADT7 using 5´ (5´-TATCGACGCT AGCGATTACA TCTATGGGAC TCAGA-3´) and 3´
(5´-AACGATCGGC GCGCCCAGCA TTAGCGTTGA GAAGTAT-3´) primers.
Fhy1 Insert2 was gained by PCR from pGADT7 using 5´ (5´-TATCGACGCT AGCAGTAGTA ATGCTGCTAA GTTTGT-3`) and 3´
(5´-AACGATCGGC GCGCCCAGCA TTAGCGTTGA GAAGTAT-3´) primers.
PhyA Insert was digested with EcoRI and SpeI, the CFP containing plasmid pAR200-cJunCFP as well. Fhy1 Insert1 was digested with AscI and NheI, the YFP containing plasmid pAR200-YFP-Dummy (for N-terminal fusion) and pAR200-cFOS-YFP (for C-terminal fusion) as well. All vector plasmids were provided by our laboratory.
Insert and Vector fragments were ligated. The purified DNA was used for a transformation in E. coli. The E. coli DNA was purified and sequenzed.
Mediums and Plates
DNA sequencing
Ligation
Plasmid spin column prep
Glycerol stocks
General Gene-Protein Information
Purification
Transformation
Dephosphorylation
Preparative Digestion
Analytic Digestion
Gel Electrophoresis
Polyacrylamide gel electrophoresis
Results
Plasmids with PhyA and Fhy1 were cloned. Unfortunately our time was running out and we only gained plasmids with mutations yet.
Discussion
The PhyA - Fhy1 light sensing system is a promising approach for regulating protein/enzyme activity by light stimulation. Of course we need evidence for the correct arrangement, given by the CFP and YFP fusion. Gaining a FRET would help us to be sure about the arrangement for split enzymes. These parts could be fused to PhyA and Fhy1 and get a complementation of the enzyme by light. In our Fusion Parts-Kit, two different split enzymes are provided (DHFR: BBa_1757005 and BBa_1757006; ß-Lactamase: BBa_1757011 and BBa_1757012). Both could be fused to PhyA and Fhy1, as well as CFP (Cerulean) and YFP (Venus).
[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Freiburg Parts]
References
Hiltbrunner A, Tscheuschler A, Viczian A, Kunkel T, Kircher S, Schäfer E. "FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor." Plant Cell Physiol. 2006 Aug;47(8):1023-34.
Hiltbrunner A, Viczian A, Bury E, Tscheuschler A, Kircher S, Toth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E "Nuclear accumulation of the phytochrome A photoreceptor requires FHY1." Curr Biol. 2005 Dec 6;15(23):2125-30.
Gambetta GA, Lagarias JC "Genetic engineering of phytochrome biosynthesis in bacteria." Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10566-71.
Takeharu Nagai, Keiji Ibata, Eun Sun Park, Mie Kubota, Katsuhiko Mikoshiba, Atsushi Miyawaki "A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications." Nature Biotechnology 20, 87 - 90 (01 Jan 2002) Research
Mark A Rizzo, Gerald H Springer, Butch Granada & David W Piston "An improved cyan fluorescent protein variant useful for FRET." Nature Biotechnology 22, 445 - 449 (01 Apr 2004) Research