Glasgow/Wetlab/Resources
From 2007.igem.org
(→Gene Sources) |
|||
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{| valign=top cellpadding=3 | {| valign=top cellpadding=3 | ||
|- | |- | ||
| - | !align=center|[[Image:Uog.jpg]] || [[Glasgow|<font face=georgia color=#3366CC size=4>Back To <br> Glasgow's <br> Main Page</font>]] || [[Glasgow/Wetlab|<font face=georgia color=#3366CC size=4>Back To <br> Glasgow's <br> Wetlab Log</font>]]|| | + | !align=center|[[Image:Uog.jpg]] || [[Glasgow|<font face=georgia color=#3366CC size=4>Back To <br> Glasgow's <br> Main Page</font>]] || [[Glasgow/Wetlab|<font face=georgia color=#3366CC size=4>Back To <br> Glasgow's <br> Wetlab Log</font>]]|| |
|} | |} | ||
---- | ---- | ||
| Line 44: | Line 44: | ||
| - | === | + | === Plasmids === |
Diagrams of the sequences from which we made our parts. | Diagrams of the sequences from which we made our parts. | ||
| Line 50: | Line 50: | ||
|- align="left" | |- align="left" | ||
| - | !width="180" |<font face=georgia color=#3366CC size=4> | + | !width="180" |<font face=georgia color=#3366CC size=4>XylR, pu and pr inpGLTUR</font> |
<br> [[Glasgow/Wetlab/References|Willardson ''et al'', 1998]] | <br> [[Glasgow/Wetlab/References|Willardson ''et al'', 1998]] | ||
| - | || [[Image:PGLTUR-2.PNG]] | + | || [[Image:PGLTUR-2.PNG|300px]] |
| + | |-align="left" | ||
| + | !width="180" |<font face=georgia color=#3366CC size=4>DntR and pDntR</font> | ||
| + | <br> [[Glasgow/Wetlab/References|Johnson ''et al'', 2002]] | ||
| + | || [[Image:DntR.PNG|300px]] | ||
| + | |||
| + | |-align="left" | ||
| + | !width="180" |<font face=georgia color=#3366CC size=4>pQF52</font> | ||
| + | <br> [[Glasgow/Wetlab/References|Tran ''et al'', 1998]] | ||
| + | || [[Image:PQF52 2.PNG]] | ||
|} | |} | ||
| Line 59: | Line 68: | ||
{| valign=top cellpadding=3 | {| valign=top cellpadding=3 | ||
|- | |- | ||
| - | !align=center|[[Image:Uog.jpg]] || || [[Glasgow|<font face=georgia color=#3366CC size=4>Back To <br> Glasgow's <br> Main Page</font>]] || [[Glasgow/Wetlab|<font face=georgia color=#3366CC size=4>Back To <br> Glasgow's <br> Wetlab Log</font>]] | + | !align=center|[[Image:Uog.jpg]] || || [[Glasgow|<font face=georgia color=#3366CC size=4>Back To <br> Glasgow's <br> Main Page</font>]] || [[Glasgow/Wetlab|<font face=georgia color=#3366CC size=4>Back To <br> Glasgow's <br> Wetlab Log</font>]] || |
|} | |} | ||
Latest revision as of 15:21, 26 October 2007
 | Back To Glasgow's Main Page | Back To Glasgow's Wetlab Log |
|---|
Resources
Some resources we have found useful in our research.
| [http://www.mbio.ncsu.edu/BioEdit/bioedit.html BioEdit] | Biological sequence alignment editor |
|---|---|
| [http://www.ncbi.nlm.nih.gov/BLAST/ BLAST] | Compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. |
| [http://www.ncbi.nlm.nih.gov/ NCBI] | National Center fot Biotechnology Information. |
| [http://openwetware.org/wiki/Main_Page Open Wetware] | DNA sequence analysis. |
| [http://www.acaclone.com/ pDRAW32] | DNA sequence analysis. |
| [http://fokker.wi.mit.edu/primer3/input.htm Primer3] | Checking primer design. |
| [http://www.iit-biotech.de/iit-cgi/oligo-tm.pl Primer Melting Temperatures] | Calculating primer melting temperatures for DNA sequencing. |
| [http://rna.lundberg.gu.se/cutter2/ Webcutter 2.0] | Restriction mapping. |
Plasmids
Diagrams of the sequences from which we made our parts.
| XylR, pu and pr inpGLTUR | 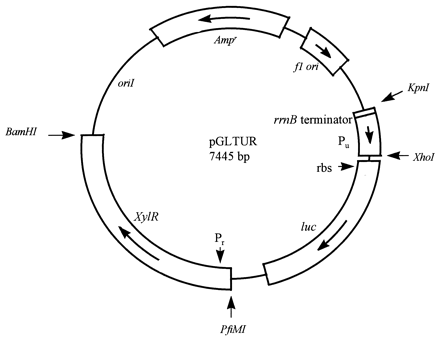
|
|---|---|
| DntR and pDntR | 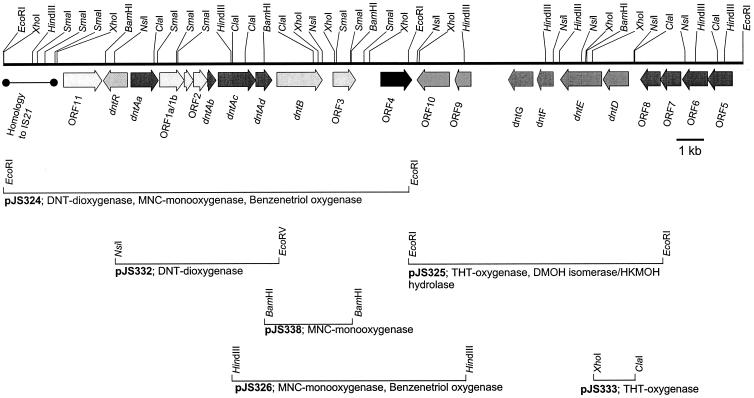
|
| pQF52 | 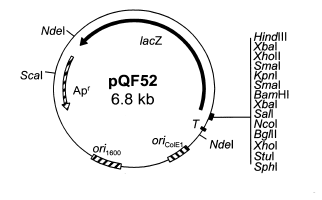
|
 | Back To Glasgow's Main Page | Back To Glasgow's Wetlab Log |
|---|