Cambridge
From 2007.igem.org
| (14 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{| cellpadding="0" cellspacing="10" | {| cellpadding="0" cellspacing="10" | ||
|- valign="middle" | |- valign="middle" | ||
| - | | style="text-align: right; padding: 0.5em; border-top: 5px solid #cccccc; border-bottom: 5px solid #cccccc;" colspan=" | + | | style="text-align: right; padding: 0.5em; border-top: 5px solid #cccccc; border-bottom: 5px solid #cccccc;" colspan="3"| |
| - | [[Image:Cambridge 2007 logo.png|left]] | + | [[Image:Cambridge 2007 logo.png|125px|left]] |
| - | <span style="font-size: 300%; font-family: Trebuchet MS, Arial, Helvetica, sans-serif">'''BOL: Bacteria OnLine'''</span | + | |
| - | <span style="font-size: | + | <span style="font-size: 300%; font-family: Trebuchet MS, Arial, Helvetica, sans-serif; line-height: 200%">'''BOL: Bacteria OnLine'''</span><br/> |
| + | <span style="font-size: 180%; font-family: Trebuchet MS, Arial, Helvetica, sans-serif; color: #333333">University of Cambridge iGEM 2007</span><br/> | ||
|- valign="top" | |- valign="top" | ||
| - | | style="border: 1px solid #003366; padding-top:0.4em; padding-left:1em; padding-right:1em; padding-bottom: 1em; background:#99ccff;"| | + | | style="border: 1px solid #003366; padding-top:0.4em; padding-left:1em; padding-right:1em; padding-bottom: 1em; background:#99ccff;" colspan="3"| |
<h2 style="margin:0; background:#003366; font-size:140%; font-weight:bold; border:1px solid #002447; text-align:left; padding:0.3em 0.4em; color: white;">Introduction</h2> | <h2 style="margin:0; background:#003366; font-size:140%; font-weight:bold; border:1px solid #002447; text-align:left; padding:0.3em 0.4em; color: white;">Introduction</h2> | ||
| - | + | [[Image:Cambridge Network diagram.png|300px|right]] | |
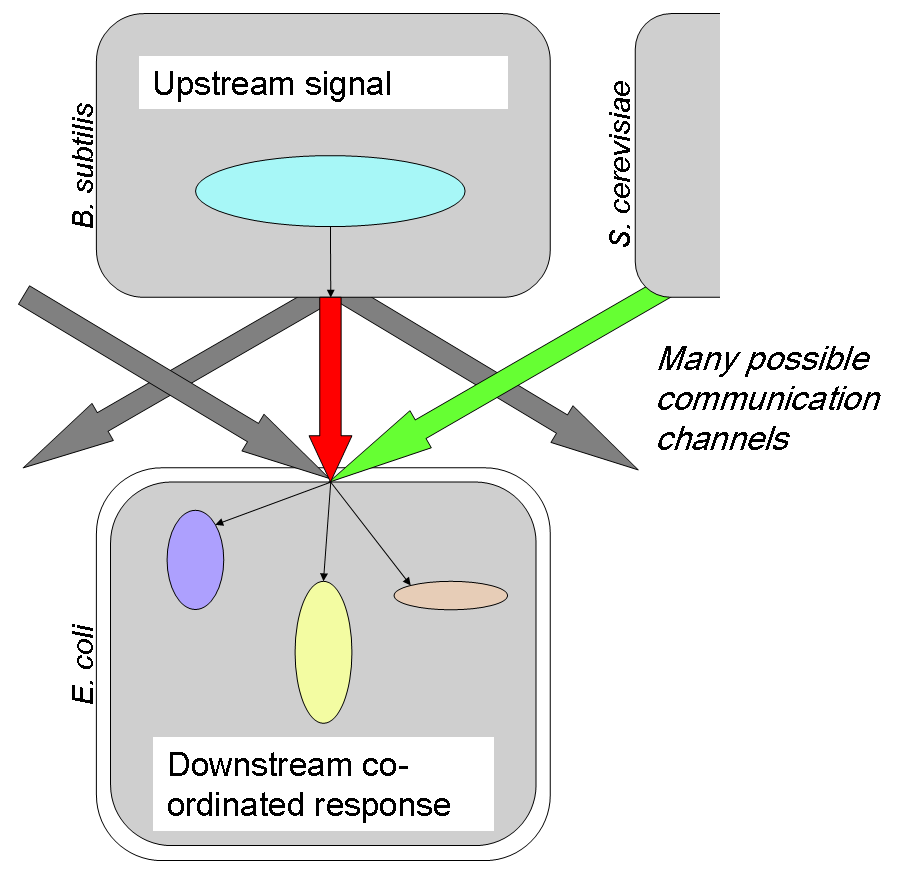
| - | + | In order to engineer interesting and useful functions in biology, a robust and extensive range of intra- and inter-cellular signalling pathways must be available. By analogy with the Internet, where adoption of the standard TCP/IP communication protocol has enabled worldwide connectivity from supercomputers to refrigerators, such a system must be accessible to cells of different heritage and structure (different “operating systems”) with the potential for processing messages received and taking action dependent on their content (see diagram at right). | |
| - | + | ||
| + | In the course of our project we identified and worked on candidates for both intracellular ([[Cambridge/Amplifier project|PoPS Amplifier project]]) and intercellular ([[Cambridge/Signalling project|Peptide signalling project]]) communication pathways, and additionally made progress towards adding a new [[Cambridge/Gram-positive chassis project|Gram-positive platform for synthetic biology]] to the Registry. | ||
| + | |||
| + | |||
| + | *[http://www.ccbi.cam.ac.uk/iGEM2007/index.php/Main_Page Go to the Cambridge iGEM 2007 team's official wiki!] (with more information about the team and our progress towards this goal) | ||
| + | |||
| + | *[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Cambridge BioBricks we have contributed to the Registry] | ||
| + | |||
| + | *Down memory lane: [[Cambridge/Snapshots of our external wiki|snapshots of our external wiki]] over the course of the project | ||
| + | |||
| + | |- width="100%" style="background: #99ccff;" cellspacing="10" valign="top" | ||
| style="border: 1px solid #003366; padding-top: 0.4em; padding-left: 1em; padding-right: 1em; background: #cce6ff;" width="33%"| | | style="border: 1px solid #003366; padding-top: 0.4em; padding-left: 1em; padding-right: 1em; background: #cce6ff;" width="33%"| | ||
| - | |||
| - | We built a standard amplifier which can be used in any synthetic transcriptional system, taking in a standard PoPS input and giving a PoPS output of a known amplification factor. In addition, the activator-promoter pairs | + | [[Image:Camb amplif icon.png|50px|left]] |
| + | |||
| + | <span style="font-weight: bold; font-size: 120%; padding: 0 0 0 0.5em; line-height: 300%;">[[Cambridge/Amplifier project|PoPS amplifier]]</span> | ||
| + | |||
| + | We BioBricked and characterised strong activators and their associated promoters. With them we built a standard amplifier which can be used in any synthetic transcriptional system, taking in a standard PoPS input and giving a PoPS output of a known amplification factor. In addition, the activator-promoter pairs studied have differential levels of activation in different combinations - this crosstalk enables their potential use for co-ordinating complex responses to stimuli. | ||
[[Cambridge/Amplifier project|More information on the amplifier project...]] | [[Cambridge/Amplifier project|More information on the amplifier project...]] | ||
| Line 26: | Line 40: | ||
| style="border: 1px solid #003366; padding-top: 0.4em; padding-left: 1em; padding-right: 0.5em; background: #cce6ff;" width="33%"| | | style="border: 1px solid #003366; padding-top: 0.4em; padding-left: 1em; padding-right: 0.5em; background: #cce6ff;" width="33%"| | ||
| - | + | [[Image:Camb agr icon.png|50px|left]] | |
| - | + | <span style="font-weight: bold; font-size: 120%; padding: 0 0 0 0.5em; line-height: 300%;">[[Cambridge/Signalling project|Peptide signalling]]</span> | |
| + | |||
| + | Peptide signalling systems exist in profusion among Gram-positive bacteria and are recognised with high specificity. For this project we chose the Agr (Accessory Gene Regulator) quorum sensing system from ''S. aureus'' as the basis of a paradigmatic peptide communication channel; our work involved implementing controllable secretion and detection of the signalling peptide AIP. In order to use such hydrophilic signal molecules in Gram-negative bacteria such as ''E. coli'', we also introduced an outer membrane pore to allow diffusion of signal molecules into the periplasm. | ||
[[Cambridge/Signalling project|More information on the signalling project...]] | [[Cambridge/Signalling project|More information on the signalling project...]] | ||
| Line 34: | Line 50: | ||
| style="border: 1px solid #003366; padding-top: 0.4em; padding-left: 1em; padding-right: 1em; background: #cce6ff;" width="33%"| | | style="border: 1px solid #003366; padding-top: 0.4em; padding-left: 1em; padding-right: 1em; background: #cce6ff;" width="33%"| | ||
| - | <span style="font-weight:bold; font-size:120%;">Gram-positive chassis</span> | + | [[Image:Camb bacillus icon.png|50px|left]] |
| + | |||
| + | <span style="font-weight: bold; font-size: 120%; padding: 0 0 0 0.5em; line-height: 300%;">[[Cambridge/Gram-positive chassis project|Gram-positive chassis]]</span> | ||
Most synthetic biology so far has involved Gram-negative bacteria, which are more widely used in molecular biology research; however, the biotechnology industry relies on Gram-positive bacteria. We worked on constructing a "chassis" out of ''Bacillus subtilis'', a common Gram-positive bacterium, into which genetic engineers can add BioBrick parts and control circuits. | Most synthetic biology so far has involved Gram-negative bacteria, which are more widely used in molecular biology research; however, the biotechnology industry relies on Gram-positive bacteria. We worked on constructing a "chassis" out of ''Bacillus subtilis'', a common Gram-positive bacterium, into which genetic engineers can add BioBrick parts and control circuits. | ||
[[Cambridge/Gram-positive chassis project|More information on the Gram-positive chassis...]] | [[Cambridge/Gram-positive chassis project|More information on the Gram-positive chassis...]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|- valign="top" | |- valign="top" | ||
| - | | style="border: 1px solid #99ccff; padding-top:0.4em; padding-left:1em; padding-right:1em; background:#cce6ff;"| | + | | style="border: 1px solid #99ccff; padding-top:0.4em; padding-left:1em; padding-right:1em; background:#cce6ff;" colspan="3"| |
<h2 style="margin:0; background:#99ccff; font-size:140%; font-weight:bold; border:1px solid #6b8fb3; text-align:left; padding:0.3em 0.4em;">Meet the team!</h2> | <h2 style="margin:0; background:#99ccff; font-size:140%; font-weight:bold; border:1px solid #6b8fb3; text-align:left; padding:0.3em 0.4em;">Meet the team!</h2> | ||
| Line 61: | Line 74: | ||
''Front row, from left:'' Narin Hengrung, Yi Han, David Wyatt | ''Front row, from left:'' Narin Hengrung, Yi Han, David Wyatt | ||
| - | ''Not in photo:'' Jim Ajioka (faculty supervisor), Lorenz Wernisch (faculty), Tony Southall ( | + | ''Not in photo:'' Jim Ajioka (faculty supervisor), Lorenz Wernisch (faculty), Jorge Goncalves (faculty supervisor), Tony Southall (postdoctoral researcher) |
The mailing list for the whole Cambridge iGEM2007 team is '''igem2007 [at] ccbi.cam.ac.uk''' | The mailing list for the whole Cambridge iGEM2007 team is '''igem2007 [at] ccbi.cam.ac.uk''' | ||
|- valign="top" | |- valign="top" | ||
| - | | style="border: 1px solid #003366; padding-top: 0.4em; padding-left: 1em; padding-right: 1em;"| | + | | style="border: 1px solid #003366; padding-top: 0.4em; padding-left: 1em; padding-right: 1em;" colspan="3"| |
<h2 style="margin:0; background:#003366; font-size:140%; font-weight:bold; border:1px solid #002447; text-align:left; padding:0.3em 0.4em; color: white;">Sponsors</h2> | <h2 style="margin:0; background:#003366; font-size:140%; font-weight:bold; border:1px solid #002447; text-align:left; padding:0.3em 0.4em; color: white;">Sponsors</h2> | ||
| Line 72: | Line 85: | ||
We are extremely grateful to the following organisations for their support of our project: | We are extremely grateful to the following organisations for their support of our project: | ||
| - | + | <span class="plainlinks">[http://www.cam.ac.uk https://static.igem.org/mediawiki/2007/9/95/Cambridge_UofC_coat_of_arms.png] [http://www.dna20.com https://static.igem.org/mediawiki/2007/1/16/Cambridge_DNA2_0_logo.png] [http://www.epsrc.ac.uk https://static.igem.org/mediawiki/2007/f/fd/Cambridge_EPSRC_logo_small.png]</span> | |
| - | + | ||
| - | + | ||
<span style="font-weight:bold; font-size:120%;">Other sponsors</span> | <span style="font-weight:bold; font-size:120%;">Other sponsors</span> | ||
| - | * The Isaac Newton Trust | + | |
| - | * | + | * [http://www.newtontrust.cam.ac.uk/ The Isaac Newton Trust UROP scheme] |
| + | * [http://www.syntheticbiology.ethz.ch/synbiocomm/ The European Union SynBioComm programme] | ||
|} | |} | ||
Latest revision as of 22:48, 26 October 2007
|
BOL: Bacteria OnLine | ||
IntroductionIn order to engineer interesting and useful functions in biology, a robust and extensive range of intra- and inter-cellular signalling pathways must be available. By analogy with the Internet, where adoption of the standard TCP/IP communication protocol has enabled worldwide connectivity from supercomputers to refrigerators, such a system must be accessible to cells of different heritage and structure (different “operating systems”) with the potential for processing messages received and taking action dependent on their content (see diagram at right). In the course of our project we identified and worked on candidates for both intracellular (PoPS Amplifier project) and intercellular (Peptide signalling project) communication pathways, and additionally made progress towards adding a new Gram-positive platform for synthetic biology to the Registry.
| ||
|
We BioBricked and characterised strong activators and their associated promoters. With them we built a standard amplifier which can be used in any synthetic transcriptional system, taking in a standard PoPS input and giving a PoPS output of a known amplification factor. In addition, the activator-promoter pairs studied have differential levels of activation in different combinations - this crosstalk enables their potential use for co-ordinating complex responses to stimuli. |
Peptide signalling systems exist in profusion among Gram-positive bacteria and are recognised with high specificity. For this project we chose the Agr (Accessory Gene Regulator) quorum sensing system from S. aureus as the basis of a paradigmatic peptide communication channel; our work involved implementing controllable secretion and detection of the signalling peptide AIP. In order to use such hydrophilic signal molecules in Gram-negative bacteria such as E. coli, we also introduced an outer membrane pore to allow diffusion of signal molecules into the periplasm. |
Most synthetic biology so far has involved Gram-negative bacteria, which are more widely used in molecular biology research; however, the biotechnology industry relies on Gram-positive bacteria. We worked on constructing a "chassis" out of Bacillus subtilis, a common Gram-positive bacterium, into which genetic engineers can add BioBrick parts and control circuits. |
Meet the team!
Top row, from left: James Brown (PhD student mentor), Jim Haseloff (faculty supervisor), Gos Micklem (faculty supervisor) Second row, from left: Yi Jin Liew, John Crowe (faculty), Lovelace Soirez, Stephanie May, Yue Miao Third row, from left: Zhizhen Zhao (Jane), Stefan Milde, Dmitry Malyshev, Xinxuan Soh (Sheila) Front row, from left: Narin Hengrung, Yi Han, David Wyatt Not in photo: Jim Ajioka (faculty supervisor), Lorenz Wernisch (faculty), Jorge Goncalves (faculty supervisor), Tony Southall (postdoctoral researcher) The mailing list for the whole Cambridge iGEM2007 team is igem2007 [at] ccbi.cam.ac.uk | ||
SponsorsWe are extremely grateful to the following organisations for their support of our project: [http://www.cam.ac.uk https://static.igem.org/mediawiki/2007/9/95/Cambridge_UofC_coat_of_arms.png] [http://www.dna20.com https://static.igem.org/mediawiki/2007/1/16/Cambridge_DNA2_0_logo.png] [http://www.epsrc.ac.uk https://static.igem.org/mediawiki/2007/f/fd/Cambridge_EPSRC_logo_small.png] Other sponsors
| ||