Edinburgh/Yoghurt
From 2007.igem.org
(→Project Goal) |
|||
| Line 22: | Line 22: | ||
* Insert biobricks into a non E. coli bacterium (Bacillus Sublillis) | * Insert biobricks into a non E. coli bacterium (Bacillus Sublillis) | ||
* Engineer a pathway to make Vanillin | * Engineer a pathway to make Vanillin | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Vanillin Project== | ==Vanillin Project== | ||
Revision as of 10:37, 8 August 2007
Edinburgh > Yoghurt
https://static.igem.org/mediawiki/2007/f/f5/800px-Edinburgh_City_15_mod.JPG
Self flavouring yoghurt
Contents |
Project Goal
Engineer lactic acid bacteria, such as Lactobacillus acidiphilus to synthesise flavours and colours during yoghurt production. The idea is to stimulate the synthesis of a certain flavour and colour via the addition of an external stimulus. Due to the nature of the finished product we will be using promoters induced by the disaccharides maltose, trehalose and arabinose.
The project will be carried out in two parts. The first is to generate the flavour and colour biosynthesis pathways in E. coli. The other part is to produce a way of placing biobricks into Lactobacillus.
Finally the flavour and colour biosynthesis pathways can be transferred to Lactobacillus.
| Targets | Operons Overview | |
|---|---|---|
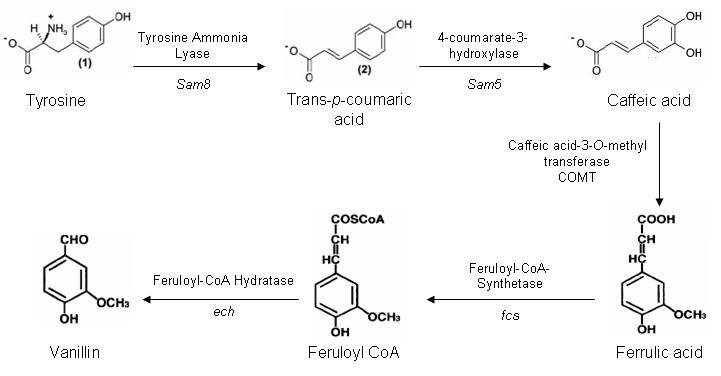
Vanillin ProjectOne of our aims for iGEM is to construct a synthetic pathway to produce vanillin from the amino acid tyrosine. There are five genes in this pathway, which are present in three different organisms. Vanillin Biosynthesis Pathway
The caffeic acid-3-0-methyltransferase gene is part of the lignin biosynthesis pathway present in plants. Our initial plan is to extranct the gene out of Alfalfa using a PCR based method. However if this method does not work we may attempt to purify COMT cDNA or have the gene artifically synthesised by GeneArt. Both fcs and ech genes are present in a variety of microorganisms. After much searching we have identified ech and fcs genes in Pseudomonas fluorescens, which have a minimal number of forbidden restriction sites. The PstI site within ech is located at the 3' terminal and shall be mutated out during the PCR amplification of the gene from P. fluorescens. Enzyme Accession NumbersSam5 and Sam8 Accession: DQ357071 Saccharothrix espanaensis culture collection: DSM 44229 COMT Accession: M63853 fcs and ech Accession: DQ119298 Pseudomonas fluorescens culture collection: NCIMB 9046 ReferencesSaccharothrix espanaensis Sam5 and Sam8 [http://jb.asm.org/cgi/content/abstract/188/7/2666 Genes and Enzymes Involved in Caffeic Acid Biosynthesis in the Actinomycete Saccharothrix esanaensis, Berner M, et al, Journal of Bacteriology 188(7): 2666, (2006)] Caffeic acid-3-O-methyltransferase mRNA Medicago sativa Gowri G, et al, Molecular cloning and expression of alfalfa S-adenosyl-L-methionine: caffeic acid 3-0-methyltransferase, a key enzyme of lignin biosynthesis, Plant Physiology 97(1):7 (1991) Downregulation of Caffeic Acid 3-O-Methyltransferase and Caffeoyl CoA 3-O-Methyltransferase in Transgenic Alfalfa Impacts on Lignin Structure and Implications for the Biosynthesis of G and S Lignin, Guo D, et al, Plant Cell. January; 13(1): 73 (2001) |