Edinburgh/Yoghurt
From 2007.igem.org
(→ech) |
(→vector) |
||
| Line 69: | Line 69: | ||
====vector==== | ====vector==== | ||
| - | * we are going to modify a vector kindly donated by Norwich University, which can act as a shuttle vector between ''E. coli'' and a broad range of gram positive hosts, including 'Bacillis subtillis'', ''Lactobacillus'' and ''Lactococcus'' | + | * we are going to modify a vector kindly donated by Norwich University, which can act as a shuttle vector between ''E. coli'' and a broad range of gram positive hosts, including ''Bacillis subtillis'', ''Lactobacillus'' and ''Lactococcus'' |
* firstly we will ligate MITs wintergreen scent generating device into the vector to characterise how the vector works in our chosen gram positive organisms | * firstly we will ligate MITs wintergreen scent generating device into the vector to characterise how the vector works in our chosen gram positive organisms | ||
Revision as of 14:12, 8 August 2007
Edinburgh > Yoghurt
https://static.igem.org/mediawiki/2007/f/f5/800px-Edinburgh_City_15_mod.JPG
Self flavouring yoghurt
Contents |
Project Discription
This project looks at applying synthetic biology techniques to Gram positive organisms for special applications. In cases such as environmental applications, vaccines, and probiotics, it would be advantageous to use alternative bacterial hosts than the traditional E.Coli and budding yeast. As a proof of concept we are attemting to engineer lactic acid bacteria, such as Lactobacillus acidiphilus to synthesise flavours during yoghurt production.
The idea is to stimulate the synthesis of a certain flavour and colour via the addition of an external stimulus. Due to the nature of the finished product we will be using promoters induced by the disaccharides maltose, trehalose and arabinose.
The project will be carried out in two parts. The first is to generate the flavour and colour biosynthesis pathways in the well characterised host bacterium E. coli. The other part is to produce a way of placing biobricks into Lactobacillus.
Finally the flavour and colour biosynthesis pathways can be transferred to Lactobacillus.
Application
There are many advantages for genetically modifying bacteria and other micro-organisms that are normally present in our food. These include increasing the number of beneficial vitamins, carotenoids and other co-factors, which may be rare in some peoples diet, such as those people suffering from malnutrition in developing countries. Other uses include the manufacture of medicines in the small intestine, which cannot survive in the gut, or biotechnological applications such as the production of naturally synthetic colours and flavours.
Vanillin Project
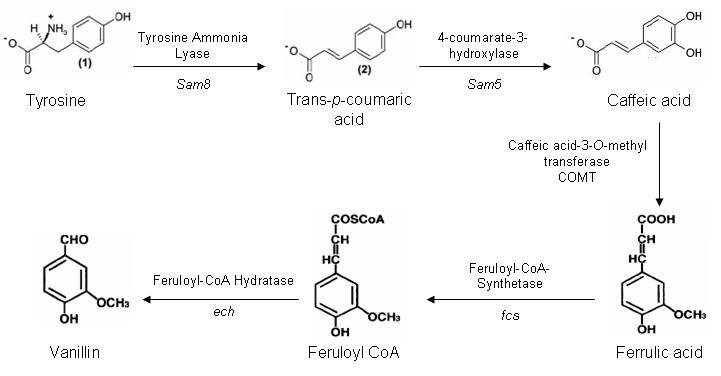
One of our aims for iGEM is to construct a synthetic pathway to produce vanillin from the amino acid tyrosine. There are five genes in this pathway, which are present in three different organisms.
Vanillin Biosynthesis Pathway
The Sam5 and Sam8 genes are from the microorganism Saccharothrix espanaensis. Sam8 encodes a tyrosine ammonia lyase enzyme, and Sam5 a hydroxylse enzyme. These genes are being kindly doanted by Anton Linnenbrink from the Institute of Pharmaceutical Sciences in Germany.
The caffeic acid-3-0-methyltransferase gene is part of the lignin biosynthesis pathway present in plants. Our initial plan is to extranct the gene out of Alfalfa using a PCR based method. However if this method does not work we may attempt to purify COMT cDNA or have the gene artifically synthesised by GeneArt.
Both fcs and ech genes are present in a variety of microorganisms. After much searching we have identified ech and fcs genes in Pseudomonas fluorescens, which have a minimal number of forbidden restriction sites. The PstI site within ech is located at the 3' terminal and shall be mutated out during the PCR amplification of the gene from P. fluorescens.
Progress so far
Sam5 & Sam8
- Saccharothrix espeanensis has arrived from DSMZ culture collection
- requires reviving and fcs and ech genes being PCR'd out
- Sam8 and Sam5 expression vector kindly donated by Dr Anton Linnenbrink from the Institute for Pharmazeutische Wissenschaften in Germany
- vector has been transformed into E. coli
- need to PCR out genes when primers arrive
COMT
- have grown up alfalfa seeds and genomic plant DNA has been extracted
- attempted to amplify up COMT via PCR, but have had no promising results so far
- RNA has been isolated from alfalfa
- have attempted to amplify up cDNA of COMT gene and are waiting on results
fcs & ech
- Pseudomonas fluorescens has arrived from NCIMB culture collection and has been revived
fcs
- have carried out several PCRs to amplify out the fcs gene from Pseudomonas fluorescenes
- PCRs did not result in one PCR product being produced, problem is most probably due to the P. fluorescens genome being of high GC content
- are attempting to purify out fcs gene from agrose gel of PCR products
ech
- the first set of primers ordered for ech did not work, as no PCR product was present after several PCR reactions
- have sent away for a second set of primers and are awaiting their arrival
vector
- we are going to modify a vector kindly donated by Norwich University, which can act as a shuttle vector between E. coli and a broad range of gram positive hosts, including Bacillis subtillis, Lactobacillus and Lactococcus
- firstly we will ligate MITs wintergreen scent generating device into the vector to characterise how the vector works in our chosen gram positive organisms
- secondly, when we have a working vanillin synthesis pathway, the vanillin biosynthesis pathway will be inserted into the vector and then transformed into Lactobacillus under the control of the arabinose promoter
- Lactobacillus may then be used to make yoghurt giving us self flavouring yoghurt, when the arabinose inducer is added
Enzyme Accession Numbers
Sam5 and Sam8 Accession: DQ357071
Saccharothrix espanaensis culture collection: DSM 44229
COMT Accession: M63853
fcs and ech Accession: DQ119298
Pseudomonas fluorescens culture collection: NCIMB 9046
References
Saccharothrix espanaensis Sam5 and Sam8
[http://jb.asm.org/cgi/content/abstract/188/7/2666 Genes and Enzymes Involved in Caffeic Acid Biosynthesis in the Actinomycete Saccharothrix esanaensis, Berner M, et al, Journal of Bacteriology 188(7): 2666, (2006)]
Caffeic acid-3-O-methyltransferase mRNA Medicago sativa
Gowri G, et al, Molecular cloning and expression of alfalfa S-adenosyl-L-methionine: caffeic acid 3-0-methyltransferase, a key enzyme of lignin biosynthesis, Plant Physiology 97(1):7 (1991)
Downregulation of Caffeic Acid 3-O-Methyltransferase and Caffeoyl CoA 3-O-Methyltransferase in Transgenic Alfalfa Impacts on Lignin Structure and Implications for the Biosynthesis of G and S Lignin, Guo D, et al, Plant Cell. January; 13(1): 73 (2001)