Tokyo/Works/Future works
From 2007.igem.org
(→アダプター) |
(→Adapters) |
||
| Line 19: | Line 19: | ||
今回の系は、全体の挙動が共存安定ある。よって、全コンストラクト | 今回の系は、全体の挙動が共存安定ある。よって、全コンストラクト | ||
| - | ==Adapters== | + | ==Adapters as a new BioBrick category== |
| - | 遺伝子回路の設計では、今回のように、ある程度コンストラクトを進めたのちにパーツの交換(TSDを使っても一度入れたパーツを取り外せるわけではないのでここは交換ではなく”新たなパーツの挿入”としたい)をしたくなるようなことが多々あるであろう。残念ながら、iGEMパーツは前後にパーツを組み合わせていくものになるので、途中のパーツの入れ替えは難しい。 | + | <!--遺伝子回路の設計では、今回のように、ある程度コンストラクトを進めたのちにパーツの交換(TSDを使っても一度入れたパーツを取り外せるわけではないのでここは交換ではなく”新たなパーツの挿入”としたい)をしたくなるようなことが多々あるであろう。残念ながら、iGEMパーツは前後にパーツを組み合わせていくものになるので、途中のパーツの入れ替えは難しい。 |
そこで我々は昨年提案したTSDstandard と合わせてアダプターという概念を提唱したい。⇒パーツページへリンク。 | そこで我々は昨年提案したTSDstandard と合わせてアダプターという概念を提唱したい。⇒パーツページへリンク。 | ||
In construction of genetic circuit, the exchange of parts after validation of circuit is required. However, current Biobrick method does not allow the insertion of new parts between two connected parts. | In construction of genetic circuit, the exchange of parts after validation of circuit is required. However, current Biobrick method does not allow the insertion of new parts between two connected parts. | ||
| - | Team Tokyo Alliance in iGEM2006 constructed TSDstandard to solve this problem. In the project, Extending the idea of Biobrick standard, together with TSDstandard that allows the insertion of parts in between, we would like to suggest the importance of Adapters. | + | Team Tokyo Alliance in iGEM2006 constructed TSDstandard to solve this problem. In the project, Extending the idea of Biobrick standard, together with TSDstandard that allows the insertion of parts in between, we would like to suggest the importance of Adapters. --> |
| + | During the construction of genetic circuits, we often find the necessity of the gene insertion between already ligated genes as in the present work. | ||
| + | |||
| + | <br>However, it is still difficult to exchange iGEM parts, which should be designed convenient for the construction, because iGEM parts have to be ligated laterally. | ||
| + | |||
| + | <br>It is necessary for more convenient BioBrick construction | ||
| + | <br>(1) to make room for parts insertion between genes after they are connected | ||
| + | <br>(2) to connect genes processed by different restriction enzymes. | ||
| + | |||
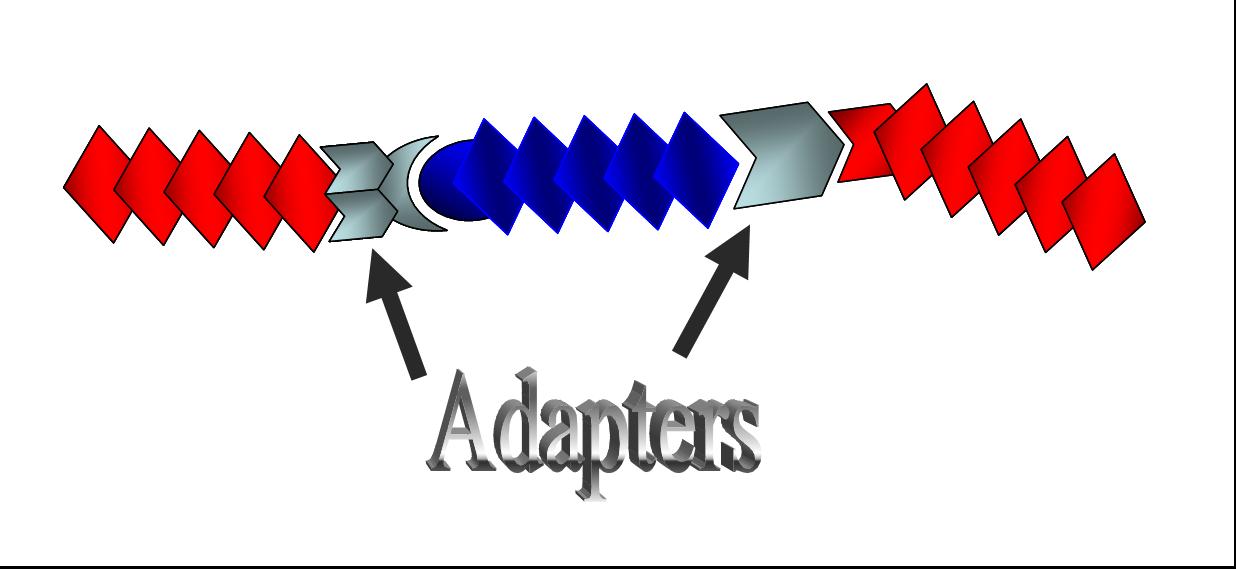
| + | <br>Here we suggest '''“adapters”''' to solve this inconvenience. In the present project, we have developed “adapters,” BioBrick parts that connect differently digested DNA, meeting the requirement (2). | ||
| + | |||
| + | <br>Together with TokyoStandard, the BioBrick parts our last year’s team Tokyo Alligance developed that enables the requirement (1), we suggest a new category of “adapter” as shown in Fig. 1. | ||
| + | |||
[[Image:adapter2.jpg|thumb|230px|'''Fig. 1 The concept of adapters''' <br>Adapters will provide more convenient Biobrick-parts-construction methods.|left]] | [[Image:adapter2.jpg|thumb|230px|'''Fig. 1 The concept of adapters''' <br>Adapters will provide more convenient Biobrick-parts-construction methods.|left]] | ||
==夢== | ==夢== | ||
Revision as of 00:33, 26 October 2007
Works top 0.Hybrid promoter 1.Formulation 2.Assay1 3.Simulation 4.Assay2 5.Future works
Balancing gene expression
According to the simulation result, we have found that the relative strength of the promoters of both sides, or relative expression levels of the downstream genes should be changed in order to reach our model. It might be possible to get closer to our goal by extremely focusing on such Wet experiments undergoing a number of trial-and-errors. However, now that we have almost obtained the strong methods of Dry approaches that compensate Wet ones, we can employ even more sophisticated methods as we have done in this project. We have estimated based on the simulation what should be done next. One option is to changing the ribosomal binding sites (RBS) to modulate largely expression level of the downstream genes. There are different RBS available as BioBrick parts, we could choose proper strength of RBS. Balancing the expression levels of both sides must be a great leap to our goal.
本アッセイへ
今回の系は、全体の挙動が共存安定ある。よって、全コンストラクト
Adapters as a new BioBrick category
During the construction of genetic circuits, we often find the necessity of the gene insertion between already ligated genes as in the present work.
However, it is still difficult to exchange iGEM parts, which should be designed convenient for the construction, because iGEM parts have to be ligated laterally.
It is necessary for more convenient BioBrick construction
(1) to make room for parts insertion between genes after they are connected
(2) to connect genes processed by different restriction enzymes.
Here we suggest “adapters” to solve this inconvenience. In the present project, we have developed “adapters,” BioBrick parts that connect differently digested DNA, meeting the requirement (2).
Together with TokyoStandard, the BioBrick parts our last year’s team Tokyo Alligance developed that enables the requirement (1), we suggest a new category of “adapter” as shown in Fig. 1.