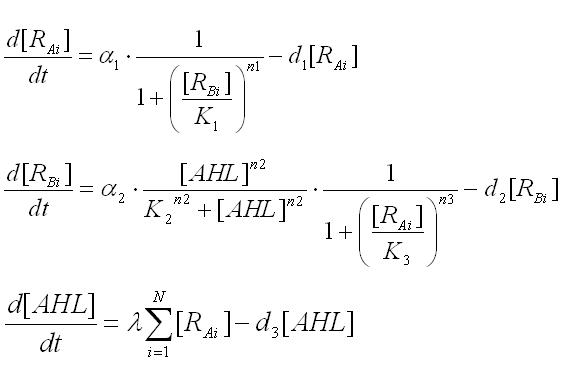Tokyo/Works/Simulation
From 2007.igem.org
| Line 20: | Line 20: | ||
<br>[[Image:expression5-1.jpg|400px|thumb|none|Ex3.2 the stochastic differential equations of the population model]] | <br>[[Image:expression5-1.jpg|400px|thumb|none|Ex3.2 the stochastic differential equations of the population model]] | ||
| - | In the simulations, Hill coefficients, coefficients of repression and activation of AHL and LacI(n2,n3,k2,k3) determined by the wet experiments were used. | + | In the simulations, Hill coefficients, coefficients of repression and activation of AHL and LacI(n2,n3,k2,k3) determined by the wet experiments were used.The values of other parameters were determined by reference to some journals. |
<br>n2 = 2.08 (-) | <br>n2 = 2.08 (-) | ||
Revision as of 01:47, 27 October 2007
Works top 0.Hybrid promoter 1.Formulation 2.Assay1 3.Simulation 4.Assay2 5.Future works
Numerical analysis and kinetic simulations for cell population
Multi-cell simulations with Hill coefficients of the promoters were carried out to find ranges of other parameters that lead the system to balanced differentiation. The differential equations for N cells were constructed to describe the cell population interacting with each other(Ex3.1).
Here the concentrations of AHL inside and outside of the cell are assumed to be the same since AHL is freely permeable though cell membranes. .
First, the phase plane analysis focused on individual cells.
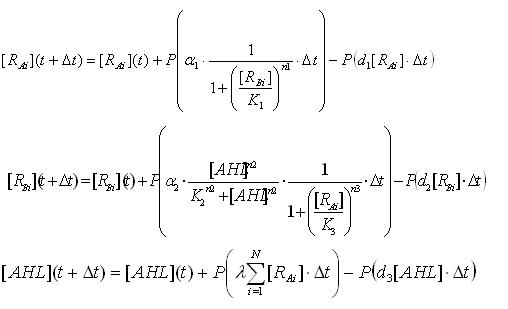
To analyze the behavior of the cell population, the above differential equations for N cells were extended to the stochastic differential equations(Ex3.2). (see detailed derivation ) In order to carry out the simulations with the stochastic model, Poisson random variables were introduced into the differential equations. As a result of the stochastic simulation, the different behaviors of individual cells were observed.
In the simulations, Hill coefficients, coefficients of repression and activation of AHL and LacI(n2,n3,k2,k3) determined by the wet experiments were used.The values of other parameters were determined by reference to some journals.
n2 = 2.08 (-)
K2 = 4.05 (μM)
n3 = 2.47 (-)
K3 = 0.295 (μM)
Determining the range of parameter which satisfy balanced differentiation
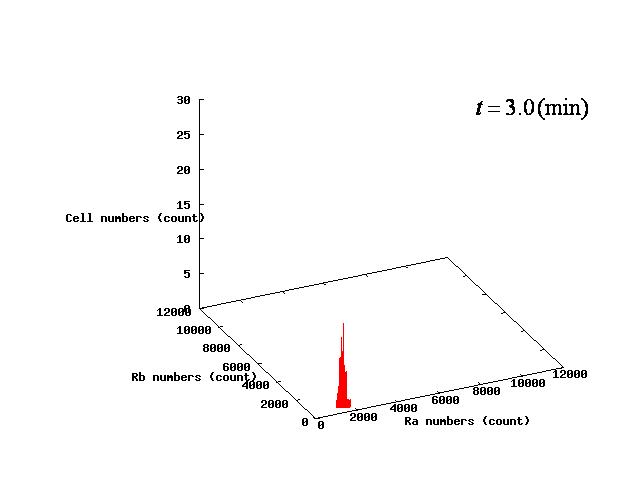
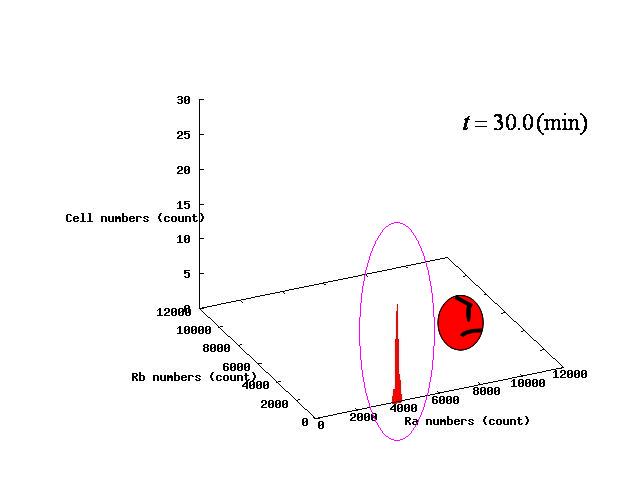
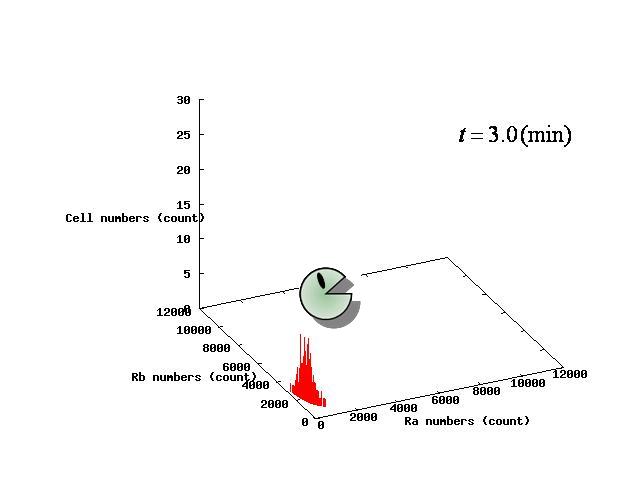
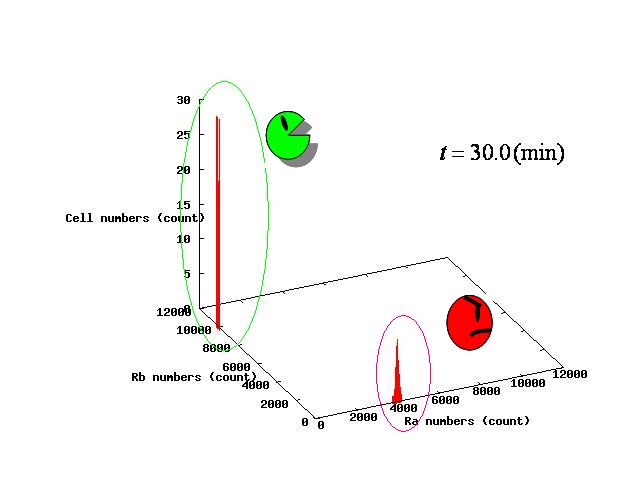
Depending on the value of parameter λ, three different patterns were observed: the pattern with coexistence state of A and B states, the pattern with only A state, and the pattern with only B. The simulation results for their patterns are shown in Fig.1 to 3.
⇒ movie here!!
⇒ movie here!!
⇒ movie here!!
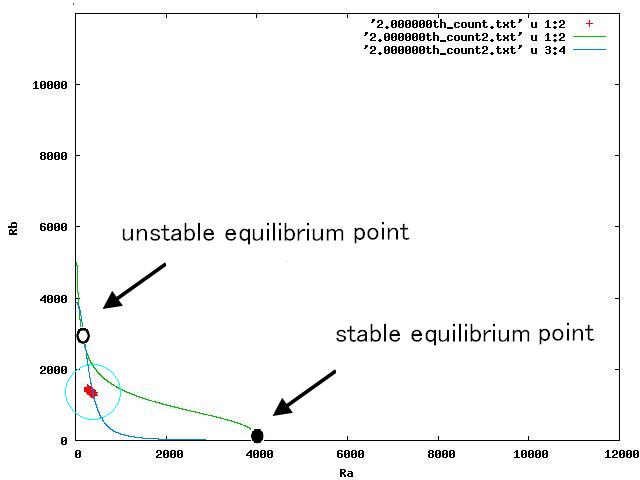
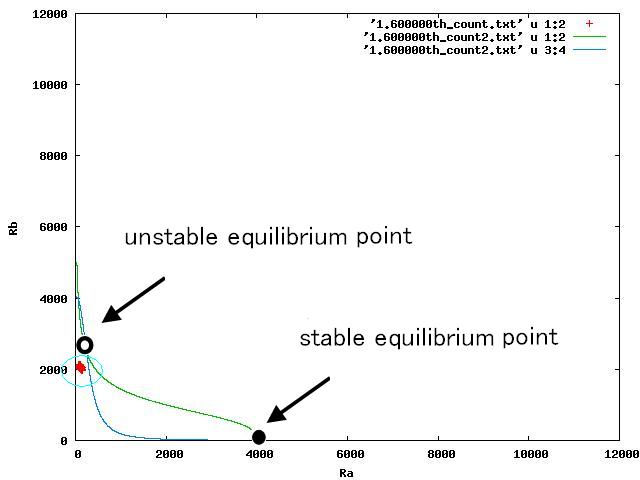
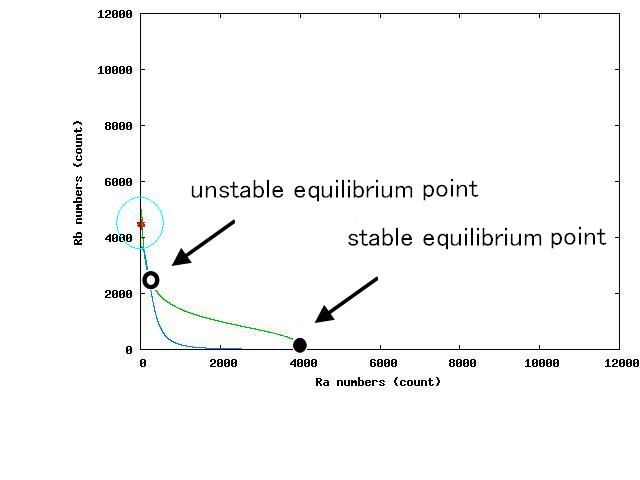
The nullclines changed dependent on the state of the cell population. When there are only B cells, the nullclines had one intersection in the phase plane. As change of the state of the cell population, the number of the intersections of nullclines turned into two. Fig.4.A-C shows the appearance of an additional intersection, indicating an unstable equilibrium point, for the three patterns, respectively.
⇒ movie about Fig.4.A here!!
⇒ movie about Fig.4.B here!!
⇒ movie about Fig.4.C here!!
In the case of Fig. 4B, the cell population distributed around the unstable equilibrium point. Right after the moment shown in the Fig. 4B, some cells moved to the A state and the other go in the direction of the B state. The division of the direction may have occurred due to the distribution of the population over the both sides of a “watershed” which is a division line lying over the unstable equilibrium point. In another possibility, fluctuation of the cell state might allow coming or going over the division line.
In the case of Fig. 4A, all the cells existed on the A-side rather than the B-side compared to the dividing line. Therefore, all the cells was not able to cross over the dividing line and moved to the A state. In contrast, when all the cells existed on the B-side rather than the A-side ( Fig 4.C), all the cells moved to the B state, similarly.
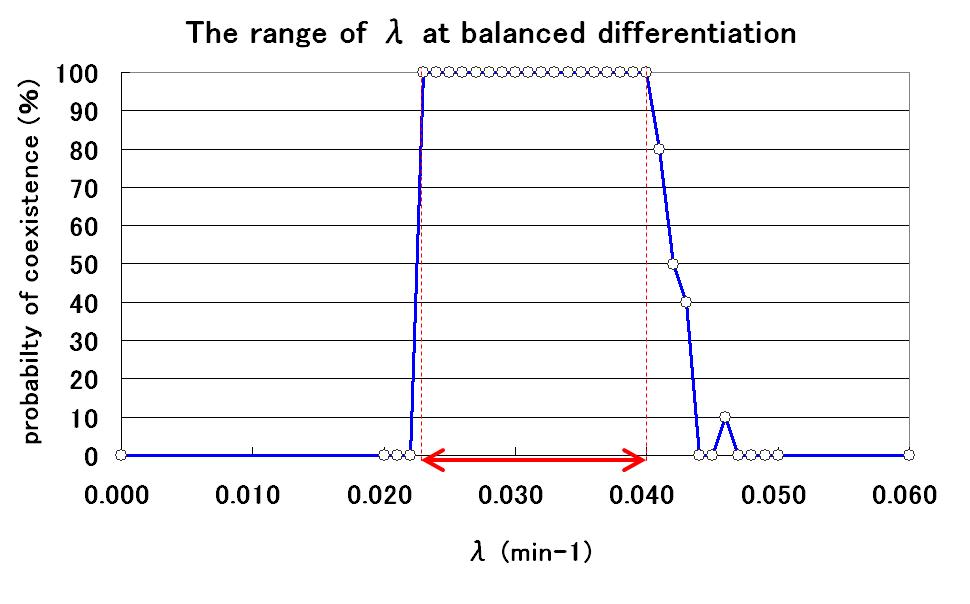
The range of parameter λ
To determine the range of the value of parameter λ for balanced differentiation, further simulations were carried out by changing the value of λ. As a result, the relationship between the value of λ and the probability of balanced differentiation was determined as shown in Fig.5.
By using parameter λ in this range, we can construct the artificial genetic circuit for balanced differentiation!