Paris/Results
From 2007.igem.org
Contents |
Experimental results
Survival of dapA deletant
If we want our system to be robust, the germ line cells should be able to live as long as possible when deprived of DAP. They may indeed face such a feat within our system if their is not enough somatic cells at a given time.
Our dapA deletant strain was grown over night in LB + 300µM DAP. A culture was then launched diluting 100 time the ON culture in LB (+ Kanamycin). 100µL were plated each hour and the number of clone determined.
dapA deletant growth
We expect the somatic cells to excrete only a limited amount of DAP, it would thus be nice if our auxotroph strain could grow on limited amount of metabolite. Over night culture were diluted 100 time in LB suplemented with different amount of DAP.
DAP excretion by prototroph cells
We want to know if prototroph cells are excreting DAP and in what amount. Instead of doing proper chemical dosages which are expensive, we had the idea to make a biological measurments.
Prototroph MG1655 strain was grown in LB, and the medium was recovered at different stages of the culture and purified. The dapA deletant strain was then grown in the recovered medium. The idea is that comparing the growth curve of this experiment with growth curves of dapA deletant in differently supplemented LB medium, should alow us to determine approimatively the amount of DAP present in the recovered medium.
Co-culture experiments
We want to know if the soma of our synthetic organism will be able to feed the germ line. Unfortunatly, this cannot properly be done without the final construct. Nevertheless, we can already check if in similar situations, dapA deleted strain can survive.
Two different co-culture experiments were performed:
- In the first one, the dapA deleted strain is grown with a prototroph strain. In this case, we know for sure that the prototroph cells will take the population over. Nervertheless it is interesting to see if dapA deleted strain survive longer in this condition than if alone and without DAP.
- In the second one, the dapA deleted strain is grown with a strain bearing an auxotrophy to another metabolite. This experiment reproduces cross-feeding works done on the yeast by (look for the exact reference). In this coculture there is a mutual dependence of each strain for the other. If this works for the dapA deleted strain with an other one, then we know for sure that dapA deleted strain can be rescued by a strain producing DAP
Recombination frequency measurements
We discussed in the design process the question of the recombination frequency. We would like to have a tunable recombination frequency in order to find the optimal one, which should be somewhere between 0 and 50% of recombination per generation.
We should be able to tune the recombination frequency by modulating the Cre recombinase expression. In order to do this, we put the Cre under the control of the pBAD promoter. We then wanted to characterize our "Cre generator device", and to make the relation between the expression level and the recombination frequency.
To do this, we used a strain (FX85, provided by Francois-Xavier Barre) having the construction lox-KmR-lox inserted into its chromosome. We first though of transforming this strain with our pBad-Cre construct (on an Ampiciline resistance plasmid), then inducing the expression with different level of arabinose, and finally spreading on selective plates (Amp or Amp+Kan). The ratio between the number of clones on the two types of plates should give us the recombination frequency. When we tried to do this, we did not get any clones on the Amp+Kan plates, even without Cre induction. But this doesn't mean that the recombination frequency is 100%. Indeed, if the recombination frequency is around 50% per generation or higher, no colonies can grow on kan plates, simply because half or more of the cells will die at each generation.
This means that our Cre generator is quite leaky and gives recombination rates too high even without induction. In order to know exactly what rate we had, we performed an other experiments. FX85 strain was transformed with our pBad-Cre construct and directly after transformation, launched into liquid culture with either Amp or Amp+Kan. 100µl of the culture were regularly plated on LB+Amp, giving the following growth curves:
dapAp characterisation
pBAD characterisation
DGAT cloning and triglycerids synthesis in E. Coli
Triglycerides are no product of the metabolism of E. coli because the bacterium miss diacylglycerol acyl-transferase (DGAT) enzyme. However the reactive necessary for the triglyceride synthesis are present in the cytoplasm: diacylglycerol is already a compound of the phospholipid catabolism (source ecocyc.org) and free fatty acids are imported from extracellular medium.
Indeed wild-type E. coli strains can grow on free fatty acid medium (oleate for example). This bacterium has Long Chain Fatty Acid (LCFA) transporter FadL. FadR, a long chain acyl-CoA-responsive transcription factor transmits the signal from FadL and controls the expression of nine genes primarily involved in fatty acid degradation and biosynthesis: it increases FadL production (so free fatty acid intake) and β-oxidation enzymes. Adding oleate in the LB medium should probably increase intake and favour the triglyceride synthesis.
Moreover qualitative model of E.coli (Palsson) has shown to perturbation in the metabolism of the bacterium. Knowing that diacylglycerol is a by-product, that free fatty acid are easily imported we have had good arguments for the success of the reaction in vivo.
We decided to use DGAT enzyme from a bacterium closely related to E. coli for metabolic compatibility. Acinetobacter calcoaceticus ADP1, Gram negative bacillus was a good candidate. Its enzyme is also an acyl-CoA fatty alcohol acyltransferase (wax ester synthase) catalyzing the final condensation of acyl-CoA and fatty alcohol. But knowing that E.coli does not produce fatty alcohol, this reaction is probably not avaible. (for more information see BBa_I718002).
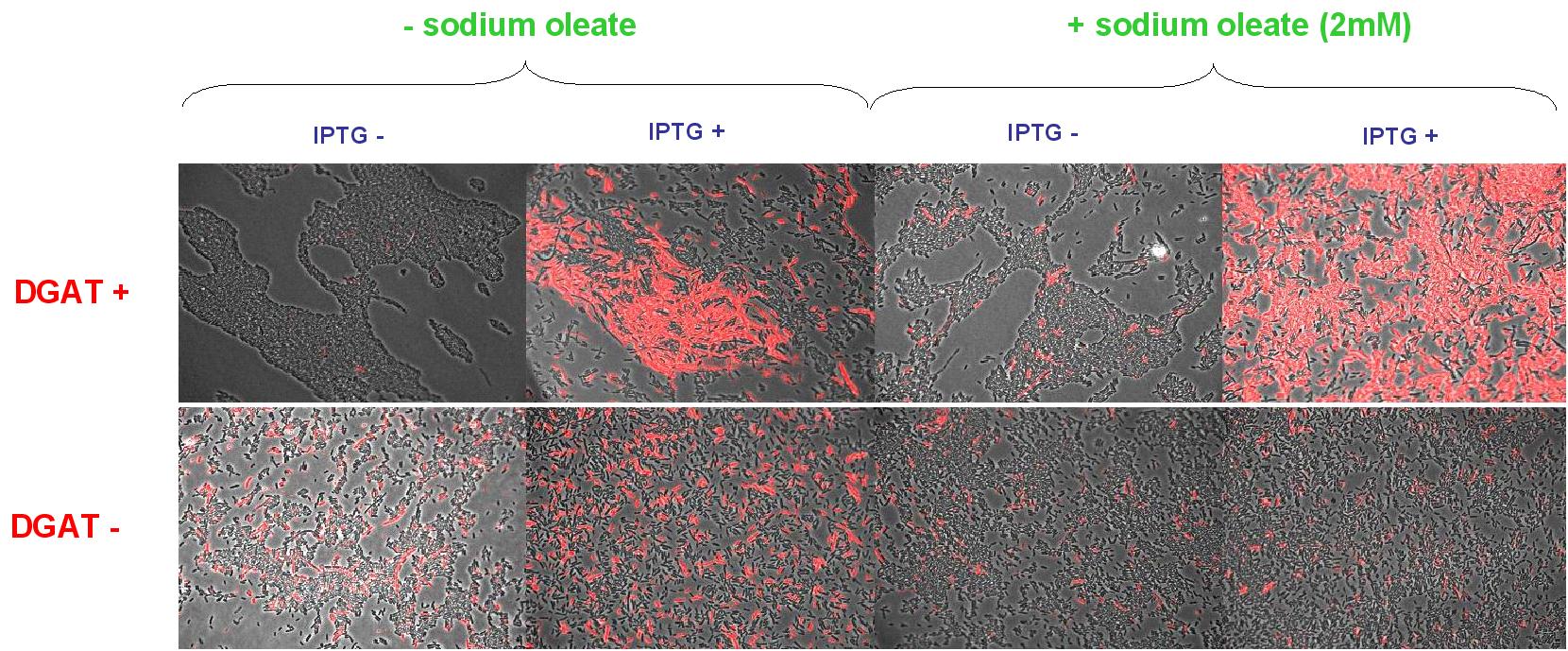
We transformed DGAT gene in pBluescript SK minus vector (Stratagene) (ampicilline resistance and pLac promoter) (pKS::DGAT) into E.coli chemically competent (DH5alpha). Gene expression is induced by IPTG. We used Nile Red fluorescence dye in the medium (5µg/mL medium) to observe lipid inclusions in different conditions (0.4mM IPTG induction in LB medium with or without sodium oleate 2mM). Results are shown below.
Line 1 represents E.coli transformed with pKS::DGAT; and Line 2 the negative control (E.coli transformed by part B0015). Columns 1 and 2 are LB medium without sodium oleate supplementation; columns 3 and 4 are LB with sodium oleate supplementation (2mM). Columns 1 and 3 are without IPTG; columns 3 and 4 with IPTG induction (0.4mM). We can observe lipid inclusion into E.coli transformed by pKS::DGAT with IPTG induction. With or without adding oleate we do not observe significant differences.
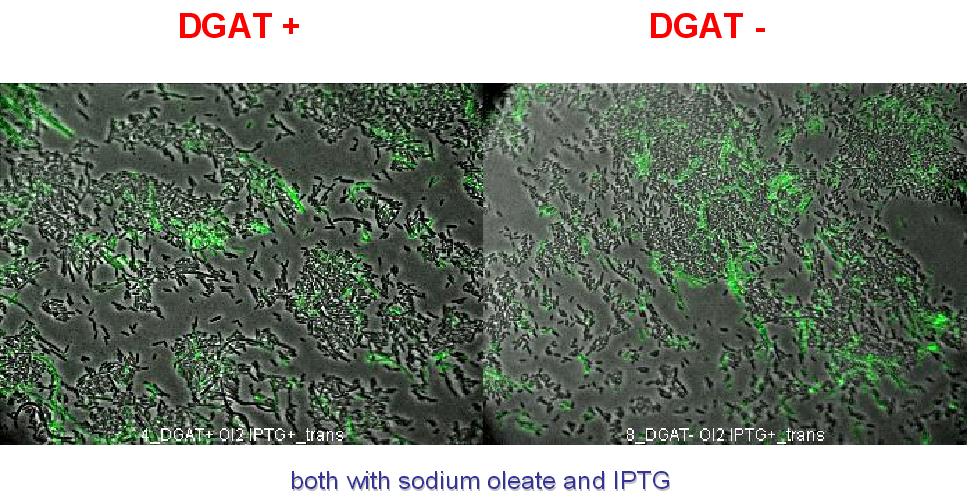
We could think that only dead cells are fluorescent (DGAT could kill the cells) but with cell death marker (green) we can see that cell death is not increased with DGAT (see below).