Edinburgh/Yoghurt/Wet Lab
From 2007.igem.org
Introduction | Applications | Objectives | Design | Modelling | Wet Lab | References
Contents |
Biobrick Creation
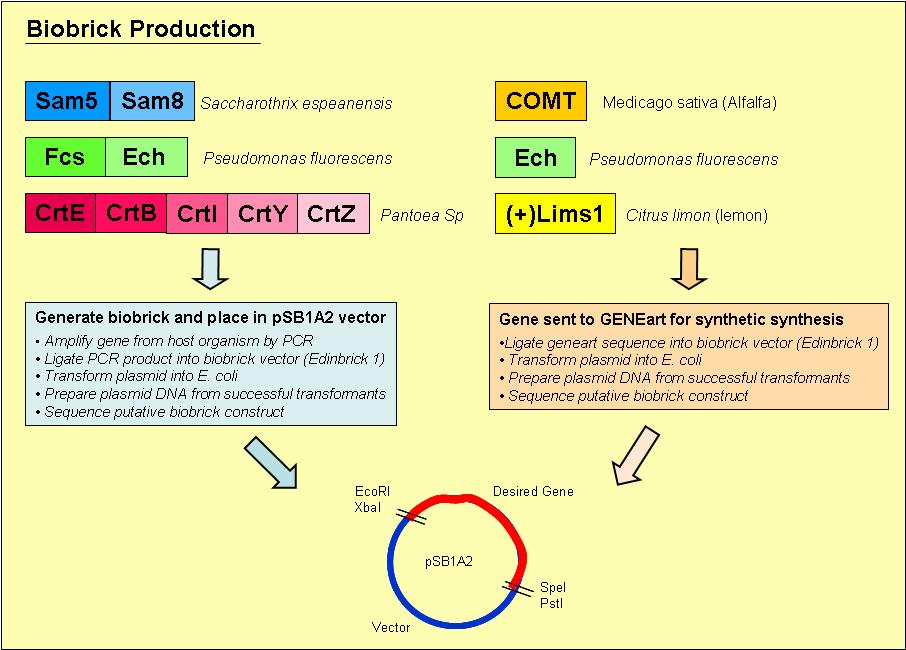
Overview of the process we used to generate our biobricks:
1. Designed primers, which included a ribosome binding site and the biobrick restriction sites, EcoRI, XbaI, PstI and SpeI, to amplify our desired gene out of its host organism
2. Purified the PCR product and loaded onto an agrose gel to check for the presence of PCR product of the right size
3. If a band was present at the right size, we digested the PCR product and vector with the restriction enzymes EcoRI & PstI
4. Digested PCR product and vector were ligated overnight with T4 DNA ligase
5. Ligated vector and gene were then transformed into E. coli and plated onto Blue/ White selection Amplicillin plates
6. Transformants that contained a product ligated into the vector would grow as white colonies and are easy to select
7. Minipreps were prepared of the white colonies
8. Purified vector DNA was digested with restriction enzymes EcoRI and PstI to determine if the vector insert was of the correct size
9. Vectors containing inserts of the correct size were sent for sequencing to confirm if they did indeed contain the correct gene
The complete methods we used to generate our biobricks may be found on the [http://openwetware.org/wiki/French_Lab French Lab] OpenWetWare site
Zeaxanthin Synthesis Pathway
Biobricks created so far
- CrtE
- CrtBI (with PstI restriction sites)
- CrtZ
Primers
Vanillin Biosynthesis Pathway
Biobricks created so far
- Sam8
Genes sent to GENEART for synthesis
- COMT
- ech
Primers
Multi Host Plasmid pTG262
So far we have modified the plasmid to contain all four biobrick restriction sites.
Introduction | Applications | Objectives | Design | Modelling | Wet Lab | References