Valencia/informational metabolism
From 2007.igem.org
Here we present a little explanation of what informational metabolism (IM) means, mainly for those who know nothing about molecular biology.
Before talking about the processes of IM, it would be interesting to take a look at the molecules involved in it:
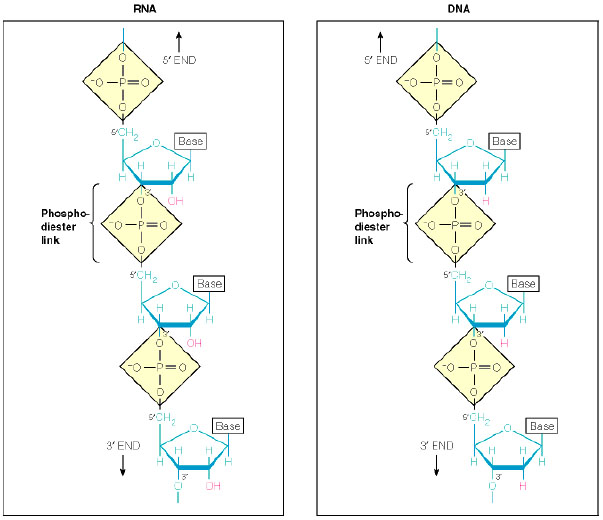
- DNA: There are two kinds of nucleid acids which are important for IM: DNA and RNA (deoxiribonucleid and ribonucleid acids) whose chemical structure can be seen on fig.3.
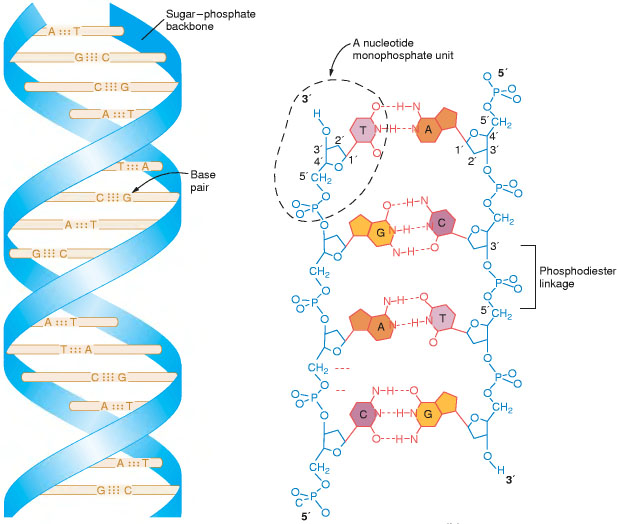
DNA is a chain of nucleotides (a sugar with a phosphate group and a base on its 5' and 1' carbon, respectively, as we show in fig.1). Nucleotides of DNA have only 4 different bases: Adenine, Guanine, Citosine and Timine. Thus, DNA can be seen as an ordered sequence of A, G, C and T.
In most organisms, DNA is present not as a linear single chain but as a doble helical chain of paired nucleotides (in such a way that A pairs only with T, and G just with C), as is shown in fig.2.
- RNA: Ribonucleic acids differ from DNA in two important items (see fig.3): the Timine base is not present, and an Uracile one (U) replaces it; and it is mostly structured in a single chain with different structures instead of a doble helical chain. There are 3 important types of RNA: mRNA, tRNA and rRNA (messenger, transfer and ribosomal respectively); in the next paragraphs we will show the task of each type in the IM.
- Proteins: Proteins are very important in metabolism, as their tasks include regulation of organisms' processes(enzimes, for instance) and structural functions (such as muscle proteins). A protein is a linear chain of aminoacids. There are 20 different aminoacids, so a protein can be seen as a sequence of ordered aminoacids. But the functionality of proteins is not due only to their aminoacid sequence; the linear chain can fold in a 3-dimensional structure, and this structure together with the aminoacid sequence are the main cause of protein functionality.
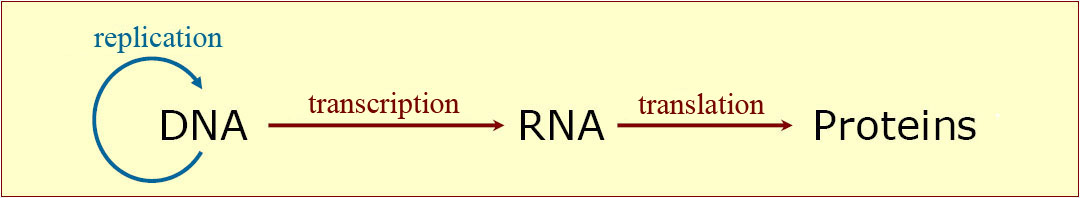
IM comprises two impotant processes in which these molecules are involved: the duplication of the hereditary genetic material (DNA) before cell division, and the production of the molecules which regulate and realize most functions of the organisms (proteins and RNA). This is sketched in fig.5.
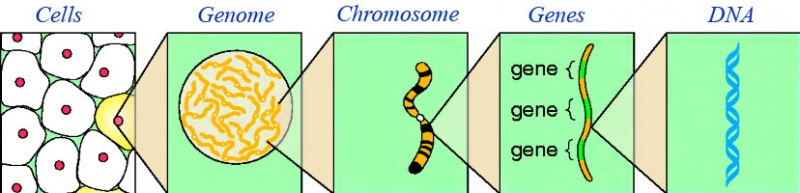
Let us, before discussing the processes of IM, have a glance to the organization of heriditary genetic material in organisms. This material is codified in DNA; the complete DNA sequence an organism has, is what we call its genome. This genome is organized inside the cells in chromosomes (for example, in E. Coli, the entire genome is condensed on a single circular chromosome, although many of them have more genetic material in little circular molecules which we call plasmids as can be seen in fig.7).
The sequence of a chromosome can be decomposed in genes. The importance of a gene is that they codify the information for at least one protein; the synthesis of proteins from genes, is what we call gene expression, and is a two step process (as we are going to see, from the DNA sequence of the gene it is generated RNA, and from this RNA, proteins are finally synthesized) that we will explain in the description of IM processes, which we tackle in the next paragraphs:
- Replication: Consists on the duplication of the cellular chromosomes before the cell division. Although we're not going to enter the details, it will be enough to mention the existence of a molecular machinery, the replicase, which controls the process: first of all, a molecule called helicase has to unwind the doble helix, and then another molecular complex called DNA-Polimerase adds to each unwinded strain, new nucleotides and bonds them. The result, is two new exact copies of the DNA doble helix wich acts as a teplate.
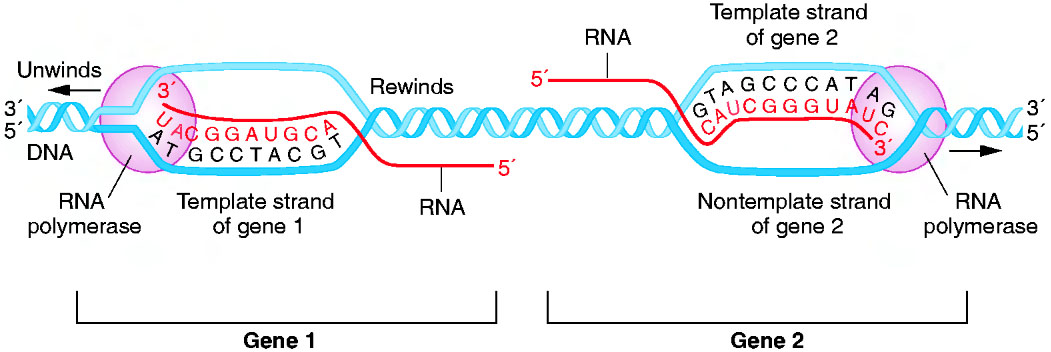
- Transcription: This is the first step of gene expression. Here, a molecular complex called RNA-Polimerase binds to a gene, unwinds DNA double helix, and uses one strand as a template for the synthesis of a RNA molecule (which will be complementary to the template DNA strand). Thus, the nucleotide sequence of this RNA molecule will be a copy of the sequence in the non-template DNA strand, but with U instead of T. This is celarly shown in fig.8.
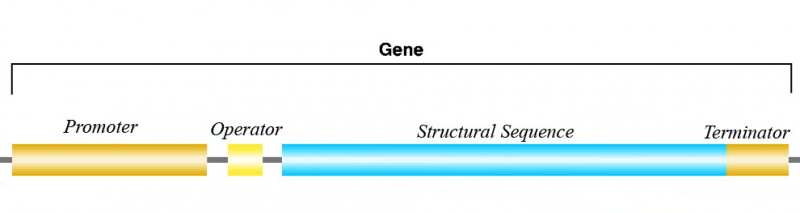
Now the question is: how know RNA-Pol where to bind in the gene, and when to finish the RNA synthesis? The answer is easy; the sequence of nucleotides a gene has, can be divided in at least 3 parts: the promoter, the structural sequence and the terminator (there is a fourth region, the operon, but this region won´t be important until we talk about gene expression regulation). So, RNA-Pol can recognize the promotor of the gene and bind to it; then, the structural sequence is transcribed as RNA until RNA-Pol arrives to the terminator, where the transcription process is stopped.
By this process all types of RNA (mRNA, tRNA and rRNA) are synthesized.
Take into acount that, as the structural sequence is the only transcribed sequence of the gene, sometimes people leaves the word gene for this structural sequence, and uses "transcription unit" to name the promotor-operator-structural-terminator united sequence. This is what we do in synthetic biology.
- Translation: Now we are going to see the final step of gene expression. Genes that codify information for the synthesis of proteins transcribe mRNA. A molecular complex called Ribosome (rich on rRNA and diverse proteins) binds to this mRNA, and synthesizes the aminoacid chain that finally will fold to form the functional protein.
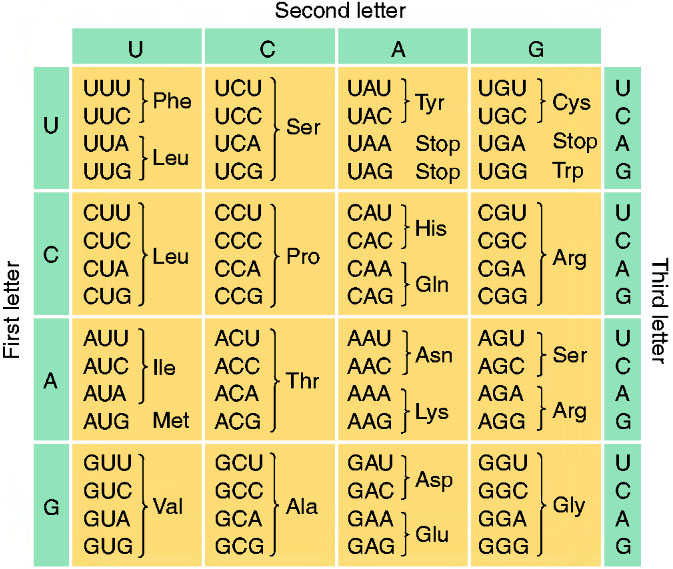
Now we have to ask one important question: how does the Ribosome read the mRNA sequence to synthesize the aminoacid chain? In nature, there are 20 different aminoacids but only 4 different bases are present in mRNA; thus, the synthesis can´t be done in a "1 nucleotide - 1 aminoacid" way. On the other hand, the synthesis of aminoacids is made in a "3 nucleotide - 1 aminoacid" way; the 3-nucleotide sequence read by the Ribosome is what we call a codon. Then, to each codon it corresponds one aminoacid, but one aminoacid can correspond to more than one codon (because of the fact that there are 4x4x4=64 different codons to obtain 20 different aminoacids), as is shown in fig.10. The AUG, UGA, UAA and UAG codons are not used in the aminoacid synthesis. When Ribosome recognize AUG, it begins to read the codons of the sequence and sinthetize the aminoacid chain, until it arrives to UGA, UAA or UAG, where the lecture is stopped (and then the protein synthesis). Then AUG serves as the initiator of the ribosomal lecture, while UGA, UAA and UAG as the terminators. Thus, finally there are 60 different codons to give 20 different aminoacids; this is what we call a degenerate code (different words have the same meaning).
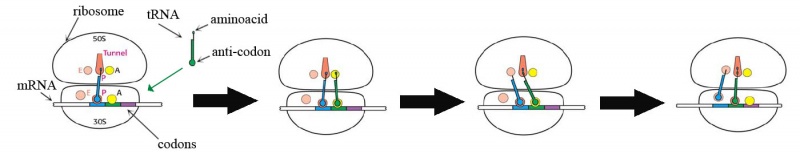
Finally, there is another question to solve, how can ribosome synthesize the aminoacid chain from the mRNA template? In this moment is when we can see the extremely important function of tRNA. This molecule (tRNA) have a very special form: in one extreme it have a 3-nucleotide sequence which we call anti-codon, and in the other extreme can assimilate the aminoacid that corresponds to the complementary codon of the anti-codon sequence. Then, as shown in fig.11, tRNA can bind to a codon of mRNA that is complementary to its anti-codon sequence, and when ribosome finds the binded mRNA-tRNA structure, it can take the aminoacid of tRNA extreme. Later, the aminoacid of the next mRNA-tRNA union can be removed from its tRNA too and added to the last aminoacid stored by ribosome. When ribosome arrives to a stop codon, the addition of aminoacids is stopped.
After all this process, the product of this process is a linear chain of aminoacids that, after a folding process, will be a protein.
This paragraphs have shown the basic processes of IM. Synthetic Biology is centered around the process of gene expression, i.e. protein synthesis. Then, a last process have to be explain:
- Gene Expression Regulation: We'd want to be able to control if a protein is expressed inside a cell, and with what rate. Obviously, we can regulate this in many ways and at each step of the expression process (transcription, traduction and folding!!). Here we are going to mention a form of regulate transcription (sketched in fig.12), as this seems to be the easier way of regulate gene expression.
Between the promoter and the structural sequence of a gene, we can find another sequence that we call the operator. For each operator, there exist a family of proteins that can bind to its sequence. This proteins, when binded to the operator site, can stop the advance of RNA-Pol (and then stop the transcription process, i.e. the RNA synthesis).In other way, this proteins can activate a non-active RNA-Pol (then, transcription cannot take place if the protein is not binded to the promoter site). This two types of regulation proteins are called repressors and activators respectively, and provides us with a mechanism for gene expression regulation.
This is the picture we find before entering on Synthetic Biology's world. Now please return to: Welcome to the world of Synthetic Biology and the iGEM competition