Davidson Missouri W
From 2007.igem.org
Wideloache (Talk | contribs) (→'''Resources / Citations''') |
Macampbell (Talk | contribs) |
||
| Line 6: | Line 6: | ||
==[[Davidson Missouri W/Project Description|Project Description]]== | ==[[Davidson Missouri W/Project Description|Project Description]]== | ||
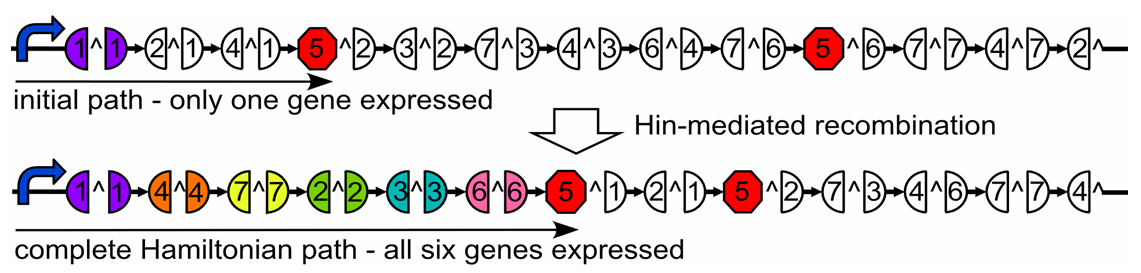
| - | The 2007 Davidson/Missouri Western iGEM team is building an E. coli computer capable of solving the Hamiltonian Path Problem. E. coli provide the massive parallel processing power required to solve this computationally intense problem from Graph Theory. We use a Hin/hixC DNA recombination mechanism to randomly generate possible paths through the graph. We then use gene expression and fragment length to screen for a Hamiltonian Path. | + | The 2007 Davidson/Missouri Western (DMW) iGEM team is building an ''E. coli'' computer capable of solving the Hamiltonian Path Problem. ''E. coli'' provide the massive parallel processing power required to solve this computationally intense problem from Graph Theory. We use a Hin/hixC DNA recombination mechanism to randomly generate possible paths through the graph. We then use gene expression and fragment length to screen for a Hamiltonian Path. |
To construct our computer, we have inserted hixC sites into reporter genes in a location that destroys the gene’s function when the split segments are not united but allows for normal gene expression when the two segments are contiguous. So far we have successfully split GFP and RFP in this manner. Our method demands that the Hin/hixC system be able to rearrange multiple DNA elements into every possible ordering. Therefore, we have tested and confirmed that Hin/hixC can rearrange 2 DNA elements into all 8 signed permutations. Our math modeling shows that it will be feasible to detect true Hamiltonian Paths even in complex graphs. | To construct our computer, we have inserted hixC sites into reporter genes in a location that destroys the gene’s function when the split segments are not united but allows for normal gene expression when the two segments are contiguous. So far we have successfully split GFP and RFP in this manner. Our method demands that the Hin/hixC system be able to rearrange multiple DNA elements into every possible ordering. Therefore, we have tested and confirmed that Hin/hixC can rearrange 2 DNA elements into all 8 signed permutations. Our math modeling shows that it will be feasible to detect true Hamiltonian Paths even in complex graphs. | ||
Revision as of 21:11, 29 August 2007
===Davidson & Missouri Western Team Logo, iGEM2007===
Project Description
The 2007 Davidson/Missouri Western (DMW) iGEM team is building an E. coli computer capable of solving the Hamiltonian Path Problem. E. coli provide the massive parallel processing power required to solve this computationally intense problem from Graph Theory. We use a Hin/hixC DNA recombination mechanism to randomly generate possible paths through the graph. We then use gene expression and fragment length to screen for a Hamiltonian Path.
To construct our computer, we have inserted hixC sites into reporter genes in a location that destroys the gene’s function when the split segments are not united but allows for normal gene expression when the two segments are contiguous. So far we have successfully split GFP and RFP in this manner. Our method demands that the Hin/hixC system be able to rearrange multiple DNA elements into every possible ordering. Therefore, we have tested and confirmed that Hin/hixC can rearrange 2 DNA elements into all 8 signed permutations. Our math modeling shows that it will be feasible to detect true Hamiltonian Paths even in complex graphs.
Team Meeting Notes
Western Meeting Notes 051407 to Present [1]
Math Related Notes
Notes 051407 to Present [2]
Students
• Will DeLoache, Junior Biology Major, [3]
• Oyinade Adefuye, Senior Biology Major, [4]
• Jim Dickson, Junior Math and Economics Major, [5]
• Amber Shoecraft, Math Major, [6]
• Andrew Martens, Senior Biology Major, [7]
• Michael Waters, Sophomore Biology Major, [8]


• Jordan Baumgardner, Junior Biology, Biochemistry/Molecular Biology Major, [9]
• Tom Crowley, Senior Biochemisty/Molecular Biology Major, [10]
• Lane H. Heard, Central High School graduate, [11]
• Nickolaus Morton, Junior Chemistry Major, [12]
• Michelle Ritter, Junior Mathematics Major, [13]
• Jessica Treece, Junior Biology Major (Health Sciences), [14]
• Matthew Unzicker, Senior Biochemistry/Molecular Biology Major, [15]
• Amanda Valencia, Senior Biochem/Molecular Biology Major, [16]
Faculty
• A. Malcolm Campbell [http://www.bio.davidson.edu/people/macampbell/macampbell.html], Professor, Department of Biology, [17]
• Karmella Haynes, Visiting Assistant Professor, Department of Biology, [18]
• [http://www.davidson.edu/math/heyer/ Laurie Heyer], Associate Professor, Department of Mathematics, [19]
Shipping Address: Malcolm Campbell, Biology Dept. Davidson College, 209 Ridge Road, Davidson, NC 28036 [(704) 894-2692]
• Todd Eckdahl [http://staff.missouriwestern.edu/~eckdahl/], Professor, Department of Biology, [20]
• Jeff Poet [http://staff.missouriwestern.edu/~poet/], Assistant Professor, Department of Computer Science, Mathematics, and Physics, [21]
Shipping Address: Todd Eckdahl, Biology Department, Missouri Western State University, 4525 Downs Drive, Saint Joseph, MO, 64507 [(816) 271-5873]
Project Overview
Hamiltonian Path Problem As a part of iGEM2006, a combined team from Davidson College and Missouri Western State University reconstituted a hin/hix DNA recombination mechanism which exists in nature in Salmonella as standard biobricks for use in E. coli. The purpose of the 2006 combined team was to provide a proof of concept for a bacterial computer in using this mechanism to solve a variation of The Pancake Problem from Computer Science. This task utilized both biology and mathematics students and faculty from the two institutions.
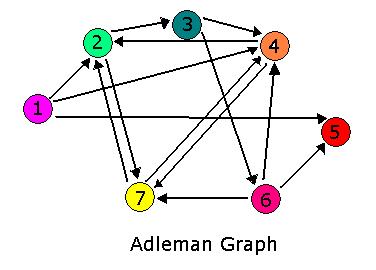
For 2007, we continue our collaboration and our efforts to manipulate E. coli into mathematics problem solvers as we refine our efforts with the hin/hix mechanism to explore another mathematics problem, the Hamiltonian Path Problem. This problem was the subject of a groundbreaking paper by Adelman in 1994 (citation below) where a unique Hamiltonian path was found in vitro for a particular directed graph on seven nodes. We propose to make progress toward solving the particular problem in vivo.
Splitting Genes for HPP
Davidson's First HPP Construct
Missouri Western's First HPP Construct
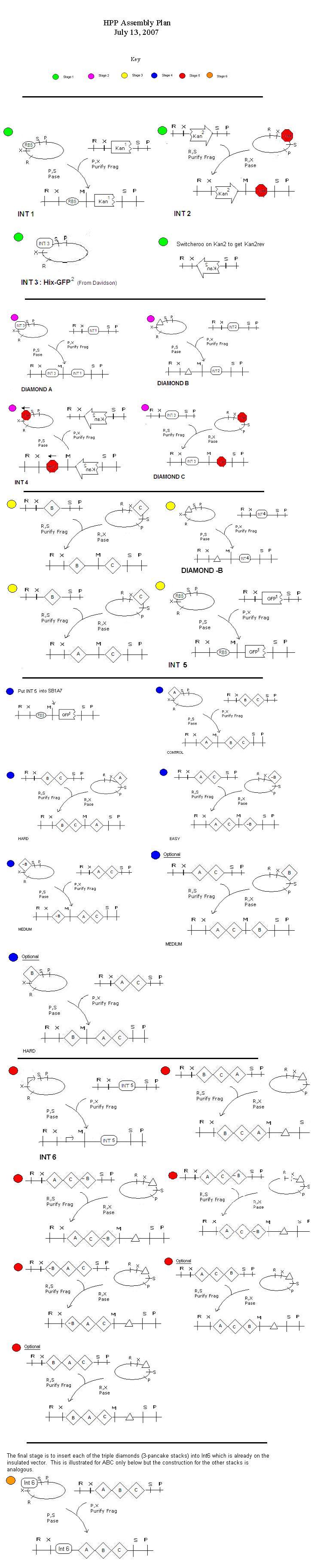
Assembly Plan
Traveling Salesperson Problem?
Synthesis of First Bacterial Computer
Resources / Citations
Missouri Western's Wet Lab Protocols
[http://gcat.davidson.edu/iGEM07/genesplitter.html Spliting Genes Web Tool]
[http://www.bio.davidson.edu/courses/Molbio/Protocols/ORIs.html Compatibility of Plasmids]
Freezer Stocks - iGEM 2006 Project
[http://spreadsheets.google.com/pub?key=pw-NamR_FPJOfhl6mDrkZcw Davidson Freezer Stocks - iGEM 2007 Project]
- DMW Part Numbers for 2007 are BBa_I715000 to BBa_I715999.
- [http://partsregistry.org/Help:BioBrick_Part_Names How to Name a New Part]
- [http://partsregistry.org/Add_a_Part_to_the_Registry Entering the Part to the Registry]
- [http://partsregistry.org/Help:Part_Features How to Annotate a Part]
Cool site for Breakfast [http://www.cut-the-knot.org/SimpleGames/Flipper.shtml]
Karen Acker's paper describing GFP and TetA(c) with Hix insertions [http://www.bio.davidson.edu/Courses/Immunology/Students/spring2006/Acker/Acker_finalpaperGFP.doc]
Bruce Henschen's paper describing one-time flippable Hix sites [http://www.bio.davidson.edu/Courses/genomics/2006/henschen/Bruce_Finalpaper.doc]
Intro to Hamiltonian Path Problem and DNA [http://www.ams.org/featurecolumn/archive/dna-abc2.html]
Adleman, LM. Molecular Computation of Solutions To Combinatorial Problems. Science. 11 November 1994. Vol. 266. no. 5187, pp. 1021 - 1024
Sambrook and Russell. 2001. Molecular Cloning A Laboratory Manual. Cold Spring Harbor Laboratry Press. Cold Spring Harbor, New York pg. 1.145. 2007 June.