Melbourne/Lab BL Notebook/20070916PCR1
From 2007.igem.org
(→Confirmation Digest) |
|||
| (21 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
=Protocol for PCR reactions A~G= | =Protocol for PCR reactions A~G= | ||
| - | For amplifying the photoreceptor and transmembrane domains of NpSopII-NpHtrII. | + | For amplifying the photoreceptor and transmembrane domains of NpSopII-NpHtrII. All -ve controls were clean. |
{| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 111: | Line 111: | ||
|} | |} | ||
| - | ==Gel Purification of PCR products A~G and 1~7== | + | ===Gel Purification of PCR products A~G and 1~7=== |
PCR product of the expected size was excised from .8% agarose gel and gel purified using the Invitrogen gel purification kit. Protocol as detailed in the kit. | PCR product of the expected size was excised from .8% agarose gel and gel purified using the Invitrogen gel purification kit. Protocol as detailed in the kit. | ||
=Second Round PCR= | =Second Round PCR= | ||
| - | For stitching products A~G to 1~7. Gel purified PCR products from the above reaction were used. | + | For stitching products A~G to 1~7. Gel purified PCR products from the above reaction were used. |
{| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 154: | Line 154: | ||
4°C forever | 4°C forever | ||
| - | |<Reaction A1, A, 1> | + | | |
| + | *<Reaction A1, A, 1> | ||
| - | <Reaction B2, B, 2> | + | *<Reaction B2, B, 2> |
| - | <Reaction C3, C, 3> | + | *<Reaction C3, C, 3> |
| - | <Reaction D4, D, 4> | + | *<Reaction D4, D, 4> |
| - | <Reaction E5, E, 5> | + | *<Reaction E5, E, 5> |
| - | <Reaction F6, F, 6> | + | *<Reaction F6, F, 6> |
| - | <Reaction G7, G, 7> | + | *<Reaction G7, G, 7> |
|- | |- | ||
| 50ul Total | | 50ul Total | ||
|} | |} | ||
| + | |||
| + | ===Gel purification of second round PCR product=== | ||
| + | All the reactions had large smears, but we cut out a band around 2.3kb (expected size) and gel purified as above. | ||
| + | |||
| + | =Digestion/Ligation of Second Round PCR product= | ||
| + | Per PCR reaction: | ||
| + | {| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | |- | ||
| + | ! Per 2° PCR {A1~G7} | ||
| + | |- | ||
| + | |6ul purified DNA {A1~G7} | ||
| + | |||
| + | 2ul 10x NEB Buffer 2 | ||
| + | |||
| + | 2ul 10x BSA | ||
| + | |||
| + | 1ul XbaI | ||
| + | |||
| + | 1ul PstI | ||
| + | |||
| + | 8ul H<sub>2</sub>O | ||
| + | |||
| + | |- | ||
| + | |||
| + | | 20ul Total | ||
| + | |} Incubated at 37°C for 45', then at RT for 6hrs. Heat inactivate at 80°C for 10'. | ||
| + | |||
| + | Also... | ||
| + | {| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | |- | ||
| + | ! BBa_J61035 (S/P) | ||
| + | |- | ||
| + | |5ul [http://partsregistry.org/Part:BBa_J61035 BBa_J61035] | ||
| + | |||
| + | 2ul 10x NEB Buffer 2 | ||
| + | |||
| + | 2ul 10x BSA | ||
| + | |||
| + | 1ul SpeI | ||
| + | |||
| + | 1ul PstI | ||
| + | |||
| + | 9ul H<sub>2</sub>O | ||
| + | |||
| + | |- | ||
| + | |||
| + | | 20ul Total | ||
| + | |} | ||
| + | |||
| + | And... | ||
| + | {| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | |- | ||
| + | ! BBa_P1010 (X/P) | ||
| + | |- | ||
| + | |5ul [http://partsregistry.org/Part:BBa_P1010 BBa_P1010](AmpR) | ||
| + | |||
| + | 2ul 10x NEB Buffer 2 | ||
| + | |||
| + | 2ul 10x BSA | ||
| + | |||
| + | 1ul XbaI | ||
| + | |||
| + | 1ul PstI | ||
| + | |||
| + | 9ul H<sub>2</sub>O | ||
| + | |||
| + | |- | ||
| + | |||
| + | | 20ul Total | ||
| + | |} | ||
| + | |||
| + | The latter two reactions were gel purified and the following ligations were set up: | ||
| + | |||
| + | ====Ligation of constructs into Death/RBS plasmid==== | ||
| + | {| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | |- | ||
| + | ! Per 2° PCR {A1~G7} | ||
| + | |- | ||
| + | |5ul X/P digested DNA {A1~G7} | ||
| + | |||
| + | 4ul X/P digested BBa_P1010 | ||
| + | |||
| + | 10ul 2x Quick ligase buffer | ||
| + | |||
| + | 1ul T4 Ligase | ||
| + | |||
| + | |- | ||
| + | |||
| + | | 20ul Total | ||
| + | |} | ||
| + | |||
| + | {| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | |- | ||
| + | ! Per 2° PCR {A1~G7} | ||
| + | |- | ||
| + | |5ul X/P digested DNA {A1~G7} | ||
| + | |||
| + | 4ul S/P digested BBa_J61035 | ||
| + | |||
| + | 10ul 2x Quick ligase buffer | ||
| + | |||
| + | 1ul T4 Ligase | ||
| + | |||
| + | |- | ||
| + | |||
| + | | 20ul Total | ||
| + | |} | ||
| + | Incubated at RT for 2hrs then [[Melbourne/Transformation_Protocol|transformed]] into competent DH5-alpha. | ||
| + | Also included some 5minute ligation incubation controls => 5 minute ligations are significantly (~5x) more efficient than 2 hours. | ||
| + | |||
| + | Pick 2 colonies per death plasmid ligation, and 4 colonies per RBS ligation and culture in LB+Amp for 12hrs. | ||
| + | |||
| + | Miniprep (nuclease free water elution) | ||
| + | |||
| + | ====Confirmation Digest==== | ||
| + | {| border="2" cellpadding="4" cellspacing="0" style="margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | |- | ||
| + | ! Per miniprepped sample | ||
| + | |- | ||
| + | |5ul miniprepped sample | ||
| + | |||
| + | 1ul 10x NEB Buffer 2 | ||
| + | |||
| + | 1ul 10x BSA | ||
| + | |||
| + | .17ul XbaI | ||
| + | |||
| + | .17ul PstI | ||
| + | |||
| + | 2.5ul H<sub>2</sub>O | ||
| + | |||
| + | |- | ||
| + | |||
| + | | 20ul Total | ||
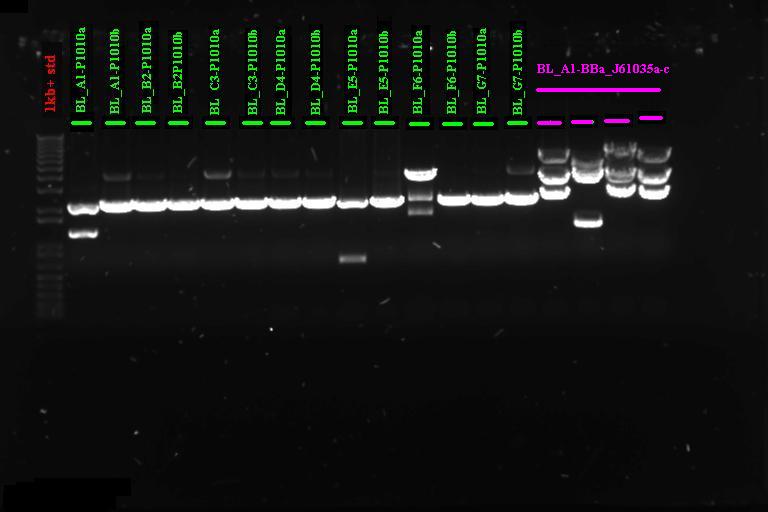
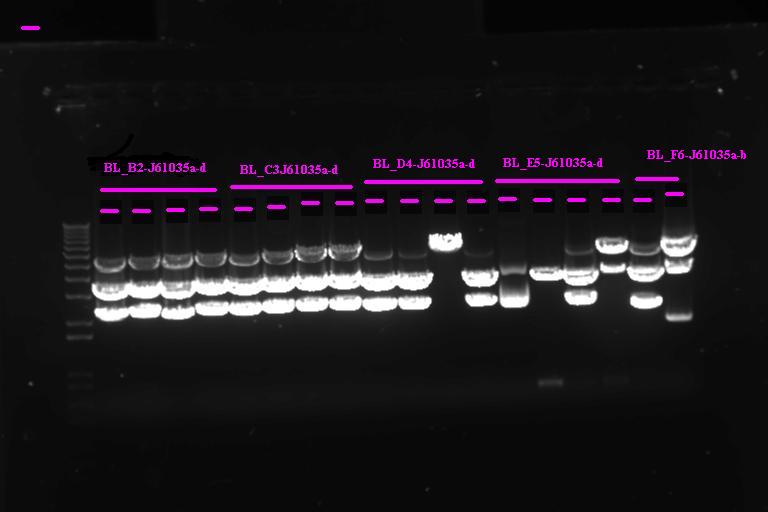
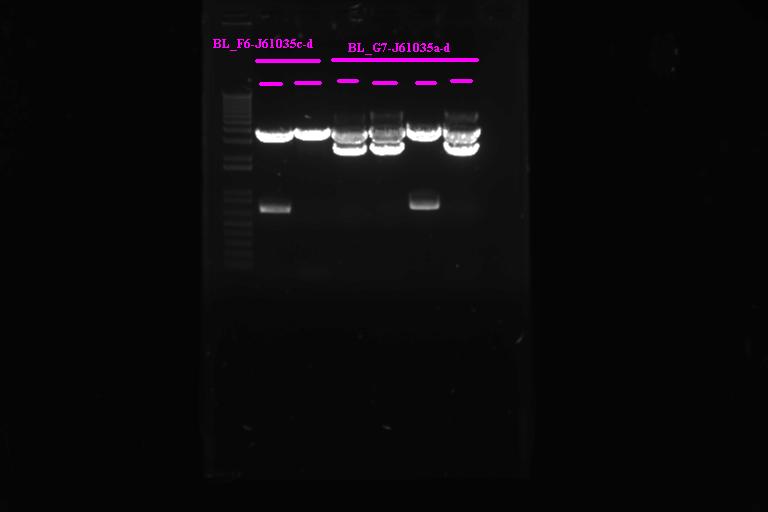
| + | |} Incubate at 37°C for 2 1hr | ||
| + | |||
| + | Run on gel | ||
| + | |||
| + | [[Image:Melbourne 20070920 bl1a.jpg|400px|Thumb|gel1]] | ||
| + | [[Image:Melbourne_Igem20070920_gel2.jpg|400px|Thumb|gel2]] | ||
| + | |||
| + | [[Image:Melbourne20070920_bl3a.jpg |400px|Thumb|gel3]] | ||
| + | |||
| + | It looks like we have the product ligated into P1010, will need to send off for sequencing. Not so sure about ligation to RBS | ||
Latest revision as of 15:19, 11 October 2007
Contents |
Protocol for PCR reactions A~G
For amplifying the photoreceptor and transmembrane domains of NpSopII-NpHtrII. All -ve controls were clean.
| PCR mix | PCR program | PrimerII |
|---|---|---|
| 5ul 10x buffer\\
5ul 10x Enhancer 0.6ul dNTPs (25mM stock) 2.5ul MgSO4 (Supplied in PCR kit) 1.5ul Primer BL_FP1_s (10uM stock) 1.5ul Primer II (10uM stock) 1ul Template (pJS010) 0.4ul Pfx Platinum (Invitrogen) 32.5ul ddH2O | 94°C - 5'
94°C - 30" 59°C - 35" 68°C - 1.5' (goto step 2 x30) 68°C -10' 4°C forever | Reaction A => Primer BL_Con1_as (Some non-specific bands)
Reaction B => Primer BL_Con2_as Reaction C => Primer BL_Con3_as (Some non-specific bands) Reaction D => Primer BL_Con4_as Reaction E => Primer BL_Con5_as Reaction F => Primer BL_Con6_as Reaction G => Primer BL_Con7_as |
| 50ul Total |
Protocol for PCR reactions 1-7
For amplification of the kinase domain of ComP
| PCR mix | PCR program | PrimerI |
|---|---|---|
| 5ul 10x buffer\\
2.5ul MgSO4 (Supplied in PCR kit) 1.5ul Primer I (10uM stock) 1.5ul Primer VR (10uM stock) 1ul Template (pJS010) 0.4ul Pfx 32.5ul ddH2O | 94°C - 5'
94°C - 30" 59°C - 35" 68°C - 1.5' (goto step 2 x30) 68°C - 10' 4°C forever | Reaction 1 => Primer BL_Con1_s
Reaction 2 => Primer BL_Con2_s Reaction 3 => Primer BL_Con3_s Reaction 4 => Primer BL_Con4_s Reaction 5 => Primer BL_Con5_s Reaction 6 => Primer BL_Con6_s Reaction 7 => Primer BL_Con7_s |
| 50ul Total |
Gel Purification of PCR products A~G and 1~7
PCR product of the expected size was excised from .8% agarose gel and gel purified using the Invitrogen gel purification kit. Protocol as detailed in the kit.
Second Round PCR
For stitching products A~G to 1~7. Gel purified PCR products from the above reaction were used.
| PCR mix | PCR program | <Reaction, TemplateI, TemplateII> |
|---|---|---|
| 5ul 10x buffer\\
2.5ul 10x Enhancer 0.6ul dNTPs (25mM stock) 2.5ul MgSO4 (Supplied in PCR kit) 1.5ul Primer BL_FP1_s (10uM stock) 1.5ul Primer VR (10uM stock) 5ul Template I {A~G} 5ul Template II {1-7} 0.4ul Pfx 27ul ddH2O | 94°C - 5'
94°C - 1' 50°C - 1' 68°C - 3' (goto step 2 x30) 68°C - 10' 4°C forever |
|
| 50ul Total |
Gel purification of second round PCR product
All the reactions had large smears, but we cut out a band around 2.3kb (expected size) and gel purified as above.
Digestion/Ligation of Second Round PCR product
Per PCR reaction:
| Per 2° PCR {A1~G7} |
|---|
| 6ul purified DNA {A1~G7}
2ul 10x NEB Buffer 2 2ul 10x BSA 1ul XbaI 1ul PstI 8ul H2O |
| 20ul Total |
Also...
| BBa_J61035 (S/P) |
|---|
| 5ul BBa_J61035
2ul 10x NEB Buffer 2 2ul 10x BSA 1ul SpeI 1ul PstI 9ul H2O |
| 20ul Total |
And...
| BBa_P1010 (X/P) |
|---|
| 5ul BBa_P1010(AmpR)
2ul 10x NEB Buffer 2 2ul 10x BSA 1ul XbaI 1ul PstI 9ul H2O |
| 20ul Total |
The latter two reactions were gel purified and the following ligations were set up:
Ligation of constructs into Death/RBS plasmid
| Per 2° PCR {A1~G7} |
|---|
| 5ul X/P digested DNA {A1~G7}
4ul X/P digested BBa_P1010 10ul 2x Quick ligase buffer 1ul T4 Ligase |
| 20ul Total |
| Per 2° PCR {A1~G7} |
|---|
| 5ul X/P digested DNA {A1~G7}
4ul S/P digested BBa_J61035 10ul 2x Quick ligase buffer 1ul T4 Ligase |
| 20ul Total |
Incubated at RT for 2hrs then transformed into competent DH5-alpha. Also included some 5minute ligation incubation controls => 5 minute ligations are significantly (~5x) more efficient than 2 hours.
Pick 2 colonies per death plasmid ligation, and 4 colonies per RBS ligation and culture in LB+Amp for 12hrs.
Miniprep (nuclease free water elution)
Confirmation Digest
| Per miniprepped sample |
|---|
| 5ul miniprepped sample
1ul 10x NEB Buffer 2 1ul 10x BSA .17ul XbaI .17ul PstI 2.5ul H2O |
| 20ul Total |
Run on gel
It looks like we have the product ligated into P1010, will need to send off for sequencing. Not so sure about ligation to RBS