Paris/July 23
From 2007.igem.org
David.bikard (Talk | contribs) |
Nicolas C. (Talk | contribs) |
||
| (21 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Paris/July 22|yesterday]] -- [[Paris/July 24|tomorrow]] <br> | ||
== w121 growth == | == w121 growth == | ||
| Line 5: | Line 6: | ||
at 12h30, the following culture was launched: | at 12h30, the following culture was launched: | ||
*w121 in 2ml LB-Erythro-DAP | *w121 in 2ml LB-Erythro-DAP | ||
| + | |||
| + | ''' | ||
| + | We were unable to perform growth kinetics analysis of w121 in different DAP-supplemented media (because the machine is occupied for the night | ||
| + | ''' | ||
== Transduction of DapA deletion in MG1655 == | == Transduction of DapA deletion in MG1655 == | ||
| Line 63: | Line 68: | ||
|- style="background: #cccccc;" | |- style="background: #cccccc;" | ||
| style="background: #ccffcc;" |D24 | | style="background: #ccffcc;" |D24 | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|eCFP bw-insert | |eCFP bw-insert | ||
|<bbpart>BBa_E0422</bbpart> (MP6.1 & MP6.2) | |<bbpart>BBa_E0422</bbpart> (MP6.1 & MP6.2) | ||
| Line 82: | Line 77: | ||
| | | | ||
|- style="background: #cccccc;" | |- style="background: #cccccc;" | ||
| - | | style="background: #ccffcc;" | | + | | style="background: #ccffcc;" |D25 |
|PoPs to GFP converter | |PoPs to GFP converter | ||
|<bbpart>BBa_E0421</bbpart> (MP7.1 & MP7.2) | |<bbpart>BBa_E0421</bbpart> (MP7.1 & MP7.2) | ||
| Line 92: | Line 87: | ||
| | | | ||
|- style="background: #cccccc;" | |- style="background: #cccccc;" | ||
| - | | style="background: #ccffcc;" | | + | | style="background: #ccffcc;" |D26 |
|gfp-tripart | |gfp-tripart | ||
|<bbpart>BBa_E0840</bbpart> (MP8.1 & MP8.2) | |<bbpart>BBa_E0840</bbpart> (MP8.1 & MP8.2) | ||
| Line 99: | Line 94: | ||
| | | | ||
|gfp-tri part (strongRBS-ORF-T) ready for use as a backward insert | |gfp-tri part (strongRBS-ORF-T) ready for use as a backward insert | ||
| + | | | ||
| + | | | ||
| + | |- style="background: #cccccc;" | ||
| + | | style="background: #ccffcc;" |D27 | ||
| + | |lox71-ftsZ bw-insert | ||
| + | |lox71-ftsZ PCR product P7 | ||
| + | |XbaI | ||
| + | |PstI | ||
| + | | | ||
| + | |lox71-ftsZ(unmutated) ready for use as a backward insert | ||
| | | | ||
| | | | ||
|} | |} | ||
| + | |||
== PCRs == | == PCRs == | ||
| + | |||
| + | {{Paris_PCR_0| Title = Lox71-FtsA-FtsZ | ||
| + | |Annealing= 60 | ||
| + | |Elongation= 3m00' | ||
| + | |Cycles= 35 | ||
| + | |Buffer= 5x 10µL | ||
| + | |MgCl2= 10µM 0µL | ||
| + | |dNTP= 10µM 1µL | ||
| + | |n_oligoF= 3 Lox71-FtsA-F | ||
| + | |v_oligoF= 2.5µL | ||
| + | |n_oligoR= 2 FtsZ-R | ||
| + | |v_oligoR= 2.5µL | ||
| + | |water= 34µl | ||
| + | |pol= Phusion 0.5µL | ||
| + | |DNA= Toothpick in MG16655 Glycerol | ||
| + | |Size= 2058 | ||
| + | |Success= | ||
| + | |Image= | ||
| + | |Band= | ||
| + | |}} | ||
| + | |||
| + | |||
| + | {{Paris_PCR_0| Title = Assembly PCR Lox71-FtsA-FtsZ-1 + FtsZ-2 | ||
| + | |Annealing= 70°C (5x) without oligos + 60°C (35x) | ||
| + | |Elongation= 3m00' | ||
| + | |Cycles= 40x | ||
| + | |Buffer= 5x 10µL | ||
| + | |MgCl2= 10µM 0µL | ||
| + | |dNTP= 10µM 1µL | ||
| + | |n_oligoF= 3 Lox71-FtsA-F | ||
| + | |v_oligoF= 2.5µL | ||
| + | |n_oligoR= 2 FtsZ-R | ||
| + | |v_oligoR= 2.5µL | ||
| + | |water= 34µl | ||
| + | |pol= Phusion 0.5µL | ||
| + | |DNA= 8.5µl Lox71-FtsA-FtsZ-1 (~50ng) + 17.5µl FtsZ-2 (~50ng) | ||
| + | |Size= 2058 | ||
| + | |Success= | ||
| + | |Image= | ||
| + | |Band= | ||
| + | |}} | ||
| + | |||
| + | |||
| + | {{Paris_PCR_0| Title = B0030-DapAColi | ||
| + | |Annealing= 60°C | ||
| + | |Elongation= 3m00' | ||
| + | |Cycles= 35x | ||
| + | |Buffer= 5x 10µL | ||
| + | |MgCl2= 10µM 0µL | ||
| + | |dNTP= 10µM 1µL | ||
| + | |n_oligoF= o20 RBS-DapAColi | ||
| + | |v_oligoF= 2.5µL | ||
| + | |n_oligoR= o7 DapAColi-R | ||
| + | |v_oligoR= 2.5µL | ||
| + | |water= 34µl | ||
| + | |pol= Phusion 0.5µL | ||
| + | |DNA= Toothpick in MG16655 Glycerol | ||
| + | |Size= 952 | ||
| + | |Success= | ||
| + | |Image= | ||
| + | |Band= | ||
| + | |}} | ||
| + | |||
| + | |||
| + | {{Paris_PCR_0| Title = B0030-DapASubtilis | ||
| + | |Annealing= 60°C | ||
| + | |Elongation= 3m00' | ||
| + | |Cycles= 35x | ||
| + | |Buffer= 5x 10µL | ||
| + | |MgCl2= 10µM 0µL | ||
| + | |dNTP= 10µM 1µL | ||
| + | |n_oligoF= o20 RBS-DapASubtilis | ||
| + | |v_oligoF= 2.5µL | ||
| + | |n_oligoR= o9 DapASubtilis-R | ||
| + | |v_oligoR= 2.5µL | ||
| + | |water= 34µl | ||
| + | |pol= Phusion 0.5µL | ||
| + | |DNA= Toothpick in MG16655 Glycerol | ||
| + | |Size= 946 | ||
| + | |Success= | ||
| + | |Image= | ||
| + | |Band= | ||
| + | |}} | ||
| + | |||
| + | == PCR mutagenesis DGAT == | ||
| + | |||
| + | DGAT has Pst1 restriction site. We want to mutagenize it to do a biobrick. | ||
| + | |||
| + | {{Paris_PCR_0| Title = DGAT1 | ||
| + | |Annealing= 50 | ||
| + | |Elongation= 2m00' | ||
| + | |Cycles= 35 | ||
| + | |Buffer= 5x 10µL | ||
| + | |MgCl2= 10µM 0µL | ||
| + | |dNTP= 10µM 1µL | ||
| + | |n_oligoF= 27 ForDGAT1 | ||
| + | |v_oligoF= 2.5µL | ||
| + | |n_oligoR= 13 DPst1-DGAT-R | ||
| + | |v_oligoR= 2.5µL | ||
| + | |water= 33µl | ||
| + | |pol= Phusion 0.5µL | ||
| + | |DNA= Miniprep pKS::DGAT (1µL = 138ng) | ||
| + | |Size= 1087 | ||
| + | |Success= YES | ||
| + | |Image= DGAT107232007.jpg | ||
| + | |Band= 1 | ||
| + | |}} | ||
| + | <br> | ||
| + | |||
| + | {{Paris_PCR_0| Title = DGAT2 | ||
| + | |Annealing= 50 | ||
| + | |Elongation= 2m00' | ||
| + | |Cycles= 35 | ||
| + | |Buffer= 5x 10µL | ||
| + | |MgCl2= 10µM 0µL | ||
| + | |dNTP= 10µM 1µL | ||
| + | |n_oligoF= 12 DPst1-DGAT | ||
| + | |v_oligoF= 2.5µL | ||
| + | |n_oligoR= 27 Rev-DGAT1 | ||
| + | |v_oligoR= 2.5µL | ||
| + | |water= 33µl | ||
| + | |pol= Phusion 0.5µL | ||
| + | |DNA= Miniprep pKS::DGAT (1µL = 138ng) | ||
| + | |Size= 396 | ||
| + | |Success= YES | ||
| + | |Image= DGAT107232007.jpg | ||
| + | |Band= 2 | ||
| + | |}} | ||
| + | <br> | ||
| + | |||
| + | == E.Coli pKS::DGAT == | ||
| + | |||
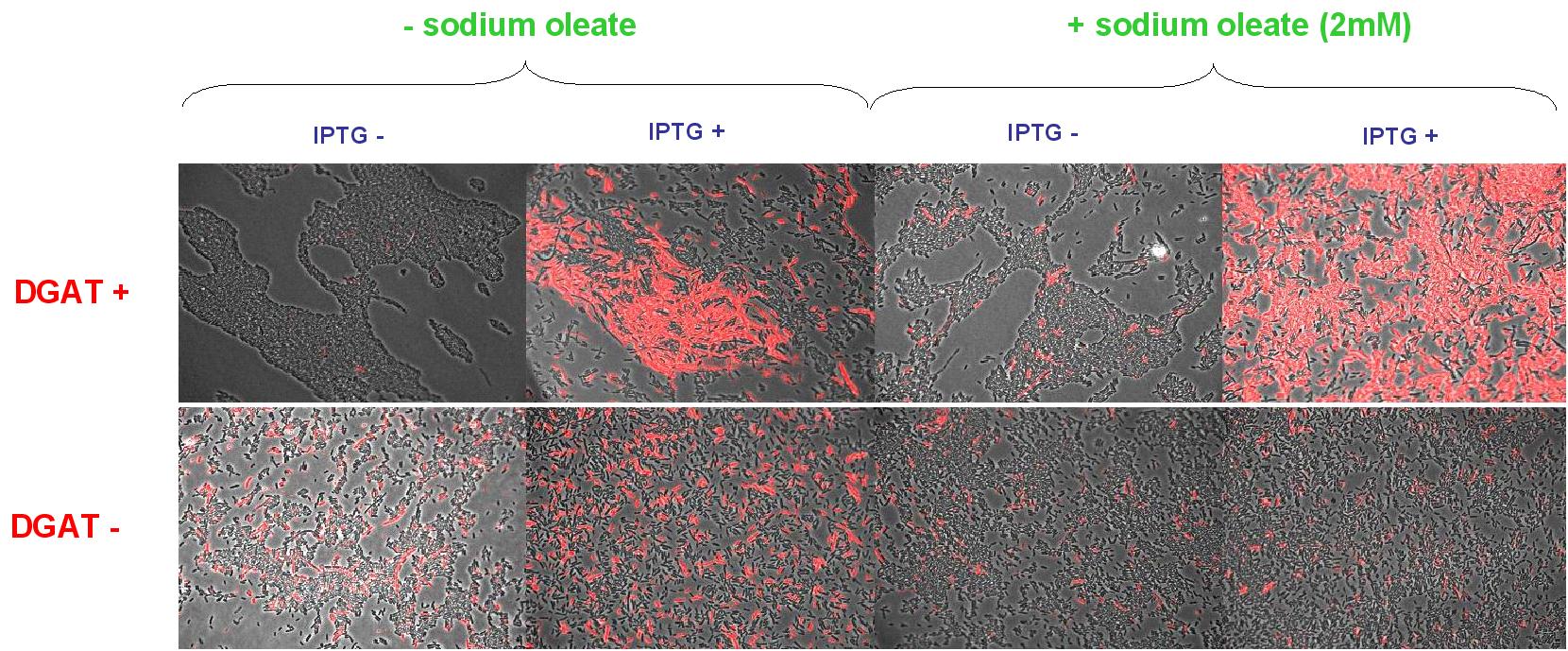
| + | We look under microscopy 5 days after incubation of E.coli transformed by pKS::DGAT and the control E.coli transformed by part B0015 on different LB medium (See [[Paris/July_18#E.coli_pKS::DGAT|July 18]]). | ||
| + | |||
| + | * Observation: | ||
| + | We can observe E.coli single cells (100X). | ||
| + | [[Image: coli_dgat_07232007.jpg|center|900px]] | ||
| + | |||
| + | |||
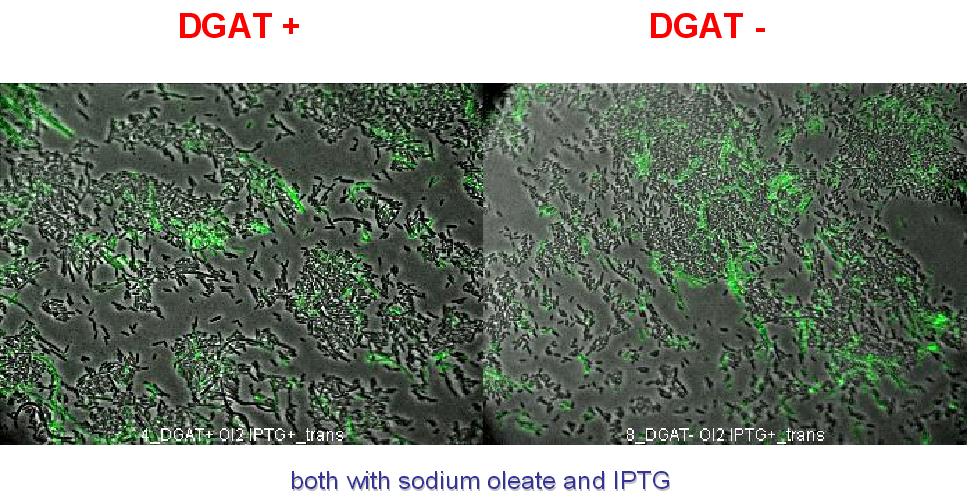
| + | * Interpretation: | ||
| + | |||
| + | We can observe lipid inclusion into E.coli transformed by pKS::DGAT with IPTG induction. With or without adding oleate we do not observe significant differences. | ||
| + | We could think that only dead cells are fluorescent (DGAT could kill the cells) but with cell death marker (green) we can see that cell death is not increased with DGAT. | ||
| + | |||
| + | [[Image: coli_dgat_death_07232007.jpg|center]] | ||
Latest revision as of 17:49, 7 October 2007
Contents |
w121 growth
w121 culture launched on 22/07/07 did not grow ON: I had forgot to add DAP to LB-Erythro growth medium.
at 12h30, the following culture was launched:
- w121 in 2ml LB-Erythro-DAP
We were unable to perform growth kinetics analysis of w121 in different DAP-supplemented media (because the machine is occupied for the night
Transduction of DapA deletion in MG1655
- The transduction seems to have worked: isolation of clones on LB and LB+DAP for verification.
Migration of digestion products
MiniPrep
Digestion reactions
Each of the digestion reactions that follow were performed as follows:
- 20µL DNA solution (miniprep or PCR product)
- 5µL Buffer 10x
- 0.5µL BSA 100x
- 2µL of each of the 2 enzymes indicated
- H20 qsp 50µL
NEB3 buffer used for XbaI/PstI double digestion NEBEcoRI buffer used for both EcoRI/SpeI & EcoRI/PstI double digestion reactions
| Digestion Products | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number | Product Name | Matrix Name | Enzyme 1 | Enzyme 2 | size | Description | ||
| D22 | pSB1A2 open vector | BBa_J61047 (MP9.1 & MP9.2) | EcoI | PstI | Open pSB1A2 vector for BioBrick EcoRI/PstI insertio | |||
| D23 | AraC/pBad promoter fw-insert | BBa_I0500 (MP4.1 & MP4.2) | EcoI | SpeI | AraC/pBad promoter ready for use as a forward insert | |||
| D24 | eCFP bw-insert | BBa_E0422 (MP6.1 & MP6.2) | XbaI | PstI | eCFP (RBS-OFR_LVA-T) ready for use as a backward insert | |||
| D25 | PoPs to GFP converter | BBa_E0421 (MP7.1 & MP7.2) | XbaI | PstI | PoPs to GFP converter ready for use as a backward insert | |||
| D26 | gfp-tripart | BBa_E0840 (MP8.1 & MP8.2) | XbaI | PstI | gfp-tri part (strongRBS-ORF-T) ready for use as a backward insert | |||
| D27 | lox71-ftsZ bw-insert | lox71-ftsZ PCR product P7 | XbaI | PstI | lox71-ftsZ(unmutated) ready for use as a backward insert | |||
PCRs
| PCR : Lox71-FtsA-FtsZ | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 2058 | |||
| 60 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 3 Lox71-FtsA-F | 2.5µL | |||
| 3m00' | Oligo R 10µM | 2 FtsZ-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
| PCR : Assembly PCR Lox71-FtsA-FtsZ-1 + FtsZ-2 | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 2058 | |||
| 70°C (5x) without oligos + 60°C (35x) | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 3 Lox71-FtsA-F | 2.5µL | |||
| 3m00' | Oligo R 10µM | 2 FtsZ-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 40x | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | 8.5µl Lox71-FtsA-FtsZ-1 (~50ng) + 17.5µl FtsZ-2 (~50ng) | |||||
| PCR : B0030-DapAColi | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 952 | |||
| 60°C | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | o20 RBS-DapAColi | 2.5µL | |||
| 3m00' | Oligo R 10µM | o7 DapAColi-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35x | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
| PCR : B0030-DapASubtilis | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 946 | |||
| 60°C | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | o20 RBS-DapASubtilis | 2.5µL | |||
| 3m00' | Oligo R 10µM | o9 DapASubtilis-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35x | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
PCR mutagenesis DGAT
DGAT has Pst1 restriction site. We want to mutagenize it to do a biobrick.
E.Coli pKS::DGAT
We look under microscopy 5 days after incubation of E.coli transformed by pKS::DGAT and the control E.coli transformed by part B0015 on different LB medium (See July 18).
- Observation:
We can observe E.coli single cells (100X).
- Interpretation:
We can observe lipid inclusion into E.coli transformed by pKS::DGAT with IPTG induction. With or without adding oleate we do not observe significant differences. We could think that only dead cells are fluorescent (DGAT could kill the cells) but with cell death marker (green) we can see that cell death is not increased with DGAT.