Samantha Liang Notebook2
From 2007.igem.org
| Line 18: | Line 18: | ||
==[[User:Samanthaliang|Samanthaliang]] 19:09, 24 July 2007 (EDT)== | ==[[User:Samanthaliang|Samanthaliang]] 19:09, 24 July 2007 (EDT)== | ||
*Subcloned 1106A.Cre and 1090.Cre (rbs.Cre) | *Subcloned 1106A.Cre and 1090.Cre (rbs.Cre) | ||
| - | *PCR with ca56(in pBad promoter)/G01001 on minipreps of 416, 417, 418, 411, 412, and | + | *PCR with ca56(in pBad promoter)/G01001 on minipreps of 416, 417, 418, 401, 411, and 412 to examine the size of the cassette - these are the ceaB variants and also a barnase and 2 bglII variants that showed some activity in the initial round of tecan screening |
| + | *Colony PCR of inactive (or rather less active) 408s - 408A, 408B, 408D, 408F | ||
| + | *The wells that I grew yesterday with the RFP reporter mechanism did not turn red at all so I'm going to examine that some more. | ||
| + | *Colony PCR of of 434 (GTG TT verision of RFP reporter) - 4 clones to look at cassette | ||
| + | *Chris saw that rbs.atg.bamH and rbs.atg.BglII wasn't lethal in Lefty, Righty, and DH10B - but will be checking if it is lethal once he adds a promoter | ||
Revision as of 23:16, 24 July 2007
My Construction Files
My Sequencing Files
My Biobricks
My -80 Stocks
First Notebook (June - July 19, 2007)
For tomorrow:
- Make large scale competent cells of Righty and Lefty
- Pick colonies if I716436 (1106A.Cre) and I716437 (1109.Cre)
- Pool I716430 through I716435 so you have minipreps of them
Samanthaliang 19:09, 24 July 2007 (EDT)
- Subcloned 1106A.Cre and 1090.Cre (rbs.Cre)
- PCR with ca56(in pBad promoter)/G01001 on minipreps of 416, 417, 418, 401, 411, and 412 to examine the size of the cassette - these are the ceaB variants and also a barnase and 2 bglII variants that showed some activity in the initial round of tecan screening
- Colony PCR of inactive (or rather less active) 408s - 408A, 408B, 408D, 408F
- The wells that I grew yesterday with the RFP reporter mechanism did not turn red at all so I'm going to examine that some more.
- Colony PCR of of 434 (GTG TT verision of RFP reporter) - 4 clones to look at cassette
- Chris saw that rbs.atg.bamH and rbs.atg.BglII wasn't lethal in Lefty, Righty, and DH10B - but will be checking if it is lethal once he adds a promoter
Samanthaliang 21:28, 23 July 2007 (EDT)
- Pick colonies from I716430 through I716435 in a 96 well block (swirl each colony in a well with arabinose and a well without) then check if the cultures turn red (might even be able to run a quick tecan on it to see the RFP levels)
- Analyzed sequencing results for 408C and 408E - unfortunately, these two guys are the same thing - [pbad][rbs][barnase - no ATG], which is not what I want because it is missing both Cre and the cassette. This must have occurred by some crazy recombination or something. I need to do some PCR mapping on some other clones to troubleshoot. I think hits are just falling out like crazy. Maybe it'd also be helpful to make libraries on low copy plasmids? Might be able to pick up more hits.
- Long google chat with Chris about next steps
Samanthaliang 15:17, 20 July 2007 (EDT)
Things to read about:
- Does barnase have DNase activity? Haven't really found anything saying that it has DNase activity yet, but it is a ribonuclease that destroys all the RNA and targets the ribosome, which in turn, stops DNA synthesis such that those bacteria should no longer be able to replicate and grow.
- Is there a way to do dapi (pronounced dap-E) measurements for e.coli? dapI binds to DNA, wonder if can use a cytometer or something? Do a dapi stain? YES! Can do a dapi stain in e.coli and then do measurements with a flow cytometer.
- Make sure that we're not just making a toxic protein or that Cre itself is toxic or something like that.
So I'm having a lot of trouble isolating the plasmid of 411B. The plate that I got from streaking a 96 well culture has really sickly single colonies that are super tiny. I can't get any of the colonies that I pick to grow in liquid media even when I add glucose. It also didn't grow after being streaked onto another plate. It seems like that in order to try to isolate a plasmid and sequence it, I'm going to have to take a swab of the plate in a dense area and miniprep that (put it in p1 buffer directly) then I'm going to have to sequence off of a PCR product.
Here's a picture of the gel:
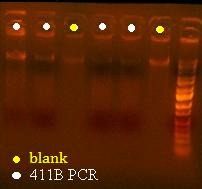
Unfortunately, there were no bands on it, meaning that either that part isn't what I think it is, or else, all the cells are already dead and the primers weren't able to pick up anything big enough to see on the gel from the plasmid. I can also see kind of a smear, which could mean that I'm picking up junk, but more likely, it's just some discoloration from the dye or something.
- Miniprep 408C, 408E, and 411B (by swabbing it)
- run PCR on 411B (ca998/G01001) and gel purify it
- Sequence 408C, 408E
- Make -80s of 408C and 408E
- Look at spotted plates - the spots actually look like they're less dense for the ones that showed a plateauing growth rate. I don't think it's obvious enough to be convincing of anything though. I guess if any cell is alive though, it would grow in the spot.