Samantha Liang Notebook2
From 2007.igem.org
| Line 14: | Line 14: | ||
*Evaluate sequencing | *Evaluate sequencing | ||
*Subclone rbs.Cre.TT with 436 and 437 to make I716438 and I716439 | *Subclone rbs.Cre.TT with 436 and 437 to make I716438 and I716439 | ||
| + | *Subclone Cre.(barnase cassette) with 212 and 240 to make I716440 and I716441 | ||
| + | *Subclone pBad.1106A with Bca1109 and Bca1106A to make I716442 | ||
Revision as of 23:40, 26 July 2007
My Construction Files
My Sequencing Files
My Biobricks
My -80 Stocks
First Notebook (June - July 19, 2007)
For tomorrow:
- Pool I716430 through I716435 so you have minipreps of them
- Evaluate sequencing
- Subclone rbs.Cre.TT with 436 and 437 to make I716438 and I716439
- Subclone Cre.(barnase cassette) with 212 and 240 to make I716440 and I716441
- Subclone pBad.1106A with Bca1109 and Bca1106A to make I716442
Contents |
Samanthaliang 19:05, 26 July 2007 (EDT)
- Colony PCR on the cultures of 408A, 408D, 411A, 434A, and 412A with and without arabinose (maybe will see some recombination and shorter PCR products for ones with arabinose that show some phenotype)
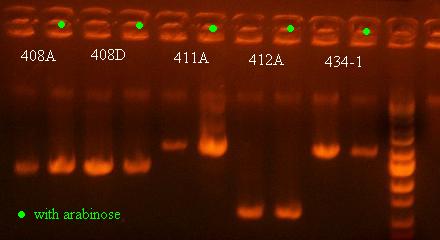
There was no change in the size of the PCR product for any of these, which I actually started to expect because the cells that are most prominent in the saturated culture will not be the cells that have undergone recombination and died already. Although it is interesting how low the bands are for 412A, which means that some recombination must have occurred for that variant as well (T.1.BglII), showing that other structures can show a recombination phenotype as well.
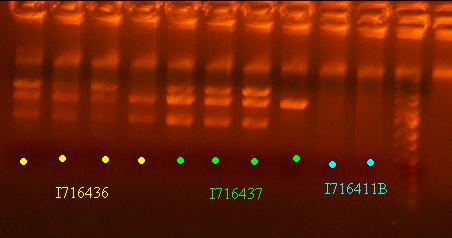
- Miniprep the 436 and 437 clones and do a quick analytic gel
- Send out 436 and 437 clones out for sequencing that pass the analytic gel test - clones 2 and 4 of I716436 and clones 1 and 2 of I716437
Samanthaliang 15:15, 25 July 2007 (EDT)
- Make large scale competent cells of Righty and Lefty
- Pick colonies if I716436 (1106A.Cre) and I716437 (1109.Cre) - 4 of each
- Grow cultures of 408A, 408D, 411A, 434A, and 412A with and without arabinose to see if there is any recombination induced by arabinose of clones that have full cassettes without being induced and yet show a slight decrease in growth from the tecan assays (under 0.95 OD at the end)
- Grow up cultures of 411B since it grew on the colony PCR plate unexpectedly. It could just be contamination or an escaped mutant but hopefully it's what I want. Also, it showed no bands on the colony PCR which is weird since it ended up growing on the plate
NOTE The new thing that i want to build is:
.pbad.rbs.atg.lox.rbs(1106A).Cre(GTG).TT.lox.barnase.
This way we can show that rbs.cre.tt is stable, then show lox.rbs.Cre.TT.lox is stable, and then focus solely on the switch by measuring the size of the cassettes and eliminate complicating studying the switch by adding toxins into the mix. After making sure the switch works by adding pbad and colony PCRing on a few clones, then add the toxin into the mix and that way we will be able to isolate the problem.
Samanthaliang 19:09, 24 July 2007 (EDT)
- Subcloned 1106A.Cre and 1090.Cre (rbs.Cre)
- PCR with ca56(in pBad promoter)/G01001 on minipreps of 416, 417, 418, 401, 411, and 412 to examine the size of the cassette - these are the ceaB variants and also a barnase and 2 bglII variants that showed some activity in the initial round of tecan screening
- Colony PCR of inactive (or rather less active) 408s - 408A, 408B, 408D, 408F
- The wells that I grew yesterday with the RFP reporter mechanism did not turn red at all so I'm going to examine that some more.
- Colony PCR of of 434 (GTG TT verision of RFP reporter) - 4 clones to look at cassette
This is annotated in my paper notebook.
- Chris saw that rbs.atg.bamH and rbs.atg.BglII wasn't lethal in Lefty, Righty, and DH10B - but will be checking if it is lethal once he adds a promoter
- Grew up cultures of Lefty and Righty
Samanthaliang 21:28, 23 July 2007 (EDT)
- Pick colonies from I716430 through I716435 in a 96 well block (swirl each colony in a well with arabinose and a well without) then check if the cultures turn red (might even be able to run a quick tecan on it to see the RFP levels)
- Analyzed sequencing results for 408C and 408E - unfortunately, these two guys are the same thing - [pbad][rbs][barnase - no ATG], which is not what I want because it is missing both Cre and the cassette. This must have occurred by some crazy recombination or something. I need to do some PCR mapping on some other clones to troubleshoot. I think hits are just falling out like crazy. Maybe it'd also be helpful to make libraries on low copy plasmids? Might be able to pick up more hits.
- Long google chat with Chris about next steps
Samanthaliang 15:17, 20 July 2007 (EDT)
Things to read about:
- Does barnase have DNase activity? Haven't really found anything saying that it has DNase activity yet, but it is a ribonuclease that destroys all the RNA and targets the ribosome, which in turn, stops DNA synthesis such that those bacteria should no longer be able to replicate and grow.
- Is there a way to do dapi (pronounced dap-E) measurements for e.coli? dapI binds to DNA, wonder if can use a cytometer or something? Do a dapi stain? YES! Can do a dapi stain in e.coli and then do measurements with a flow cytometer.
- Make sure that we're not just making a toxic protein or that Cre itself is toxic or something like that.
So I'm having a lot of trouble isolating the plasmid of 411B. The plate that I got from streaking a 96 well culture has really sickly single colonies that are super tiny. I can't get any of the colonies that I pick to grow in liquid media even when I add glucose. It also didn't grow after being streaked onto another plate. It seems like that in order to try to isolate a plasmid and sequence it, I'm going to have to take a swab of the plate in a dense area and miniprep that (put it in p1 buffer directly) then I'm going to have to sequence off of a PCR product.
Here's a picture of the gel:
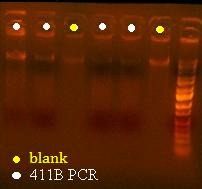
Unfortunately, there were no bands on it, meaning that either that part isn't what I think it is, or else, all the cells are already dead and the primers weren't able to pick up anything big enough to see on the gel from the plasmid. I can also see kind of a smear, which could mean that I'm picking up junk, but more likely, it's just some discoloration from the dye or something.
- Miniprep 408C, 408E, and 411B (by swabbing it)
- run PCR on 411B (ca998/G01001) and gel purify it
- Sequence 408C, 408E
- Make -80s of 408C and 408E
- Look at spotted plates - the spots actually look like they're less dense for the ones that showed a plateauing growth rate. I don't think it's obvious enough to be convincing of anything though. I guess if any cell is alive though, it would grow in the spot.