Tokyo/Works/Future works
From 2007.igem.org
(→夢) |
(→Adapters as a new BioBrick category) |
||
| (53 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| - | <br>[[Tokyo/Works|Works top]] | + | <br>[[Tokyo/Works|Works top]] 0.[[Tokyo/Works/Hybrid promoter|Hybrid promoter]] 1.[[Tokyo/Works/Formulation |Formulation]] 2.[[Tokyo/Works/Assay |Assay1]] 3.[[Tokyo/Works/Simulation |Simulation]] 4.[[Tokyo/Works/Assay2 |Assay2]] 5.[[Tokyo/Works/Future works |Future works]] |
== Balancing gene expression == | == Balancing gene expression == | ||
| Line 10: | Line 10: | ||
具体的には、プロモータのRBSを変えるのがよいだろう。これにより活性を大きく変えることができる。(具体的数値も入れた方がよい?) | 具体的には、プロモータのRBSを変えるのがよいだろう。これにより活性を大きく変えることができる。(具体的数値も入れた方がよい?) | ||
その結果からさらに調整をしていけば、必ずや目標へとたどり着けるであろう。--> | その結果からさらに調整をしていけば、必ずや目標へとたどり着けるであろう。--> | ||
| - | According to the simulation results, we have found that the relative strength of the promoters of both sides, or relative expression levels of the downstream genes should be changed in order to reach our model. | + | According to the simulation results and wet experiments, we have found that the relative strength of the promoters of both sides, or relative expression levels of the downstream genes should be changed in order to reach our model. |
| - | + | Even without Dry approaches, it might be possible to get closer to our goal by extremely focusing on such Wet experiments undergoing a number of trial-and-errors. | |
However, now that we have almost obtained the strong methods of Dry approaches that compensate Wet ones, we can employ even more sophisticated methods as we have done in this project. | However, now that we have almost obtained the strong methods of Dry approaches that compensate Wet ones, we can employ even more sophisticated methods as we have done in this project. | ||
| - | We have estimated based on the | + | <br>We have estimated, based on the simulations and the wet experiments, what should be done next. One option is to change the ribosomal binding sites (RBS) to modulate largely expression level of the downstream genes. There are different RBS available as BioBrick parts, we could choose proper strength of RBS. Another option is that change of -35 and/or -10 sequence of the promoters. Balancing the expression levels of both sides must be a great leap to our goal. |
| - | Balancing the expression levels of both sides must be a great leap to our goal. | + | |
| + | == Vice Versa== | ||
| + | <!--In the present model, only B de and redifferentiates in the absence of A but not vice versa. | ||
| + | Establishing this model, we plan to program A by signalling molecules orthogonal to AHL to dedifferentiate in the absence of B and then redifferentiate into B. | ||
| + | When the ratio of A and B reaches to a certain value, the system is balanced again.--> | ||
| + | In the present model, only B de- and redifferentiates in the absence of A but not vice versa. | ||
| + | Establishing complete balanced differentiation model, we have a plan of an improved model where A also dedifferentiates in the absence of B and then redifferentiates into B. | ||
| + | To de- and redifferentiate A, we are planning to use another signaling molecule orthogonal to the AHL for LuxR. | ||
| + | When the ratio of A and B reaches to a certain value, the system is balanced again. | ||
| + | <br><br> | ||
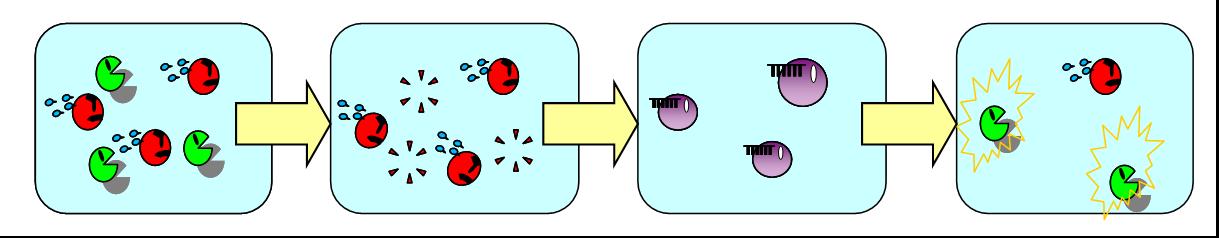
| + | [[Image:vice versa.jpg|thumb|550px|Fig. 1 Our next model where A de- and redifferentiates in the absence of B.|left]] | ||
| + | <br><br> | ||
| + | |||
| + | <br><br><br><br><br> | ||
| + | |||
| + | |||
| + | |||
| + | <br> | ||
== Single cell analysis with a cell sorter == | == Single cell analysis with a cell sorter == | ||
| - | Throughout the Wet experiments so far, we have observed our genetically engineered cells, not at an individual levels but at a collective level. With a single cell analysis, our simulation results would be more clearly confirmed. | + | Throughout the Wet experiments so far, we have observed promoter activity of our genetically engineered cells, not at an individual levels but at a collective level. With a single cell analysis, our simulation results would be more clearly confirmed. |
| - | <br>We could do this at the next step by using a cell sorter, which separates a mixture of cells into individuals. Then we could observe which states, A, B, or | + | <br>We could do this at the next step by using a cell sorter, which separates a mixture of cells into individuals. Then we could observe which states, A, B, or dedifferentiated, individual cells are taking at the moment of the measurement and how many of them are in each state. |
<!--Our model in this project aims at "coexistence stability." --> | <!--Our model in this project aims at "coexistence stability." --> | ||
| Line 30: | Line 47: | ||
Team Tokyo Alliance in iGEM2006 constructed TSDstandard to solve this problem. In the project, Extending the idea of Biobrick standard, together with TSDstandard that allows the insertion of parts in between, we would like to suggest the importance of Adapters. --> | Team Tokyo Alliance in iGEM2006 constructed TSDstandard to solve this problem. In the project, Extending the idea of Biobrick standard, together with TSDstandard that allows the insertion of parts in between, we would like to suggest the importance of Adapters. --> | ||
During the construction of genetic circuits, we often found the necessity of the gene insertion between already ligated genes as in the present work. | During the construction of genetic circuits, we often found the necessity of the gene insertion between already ligated genes as in the present work. | ||
| - | |||
<br>However, it is still difficult to exchange iGEM parts, which should be designed convenient for the construction, because iGEM parts have to be ligated laterally. | <br>However, it is still difficult to exchange iGEM parts, which should be designed convenient for the construction, because iGEM parts have to be ligated laterally. | ||
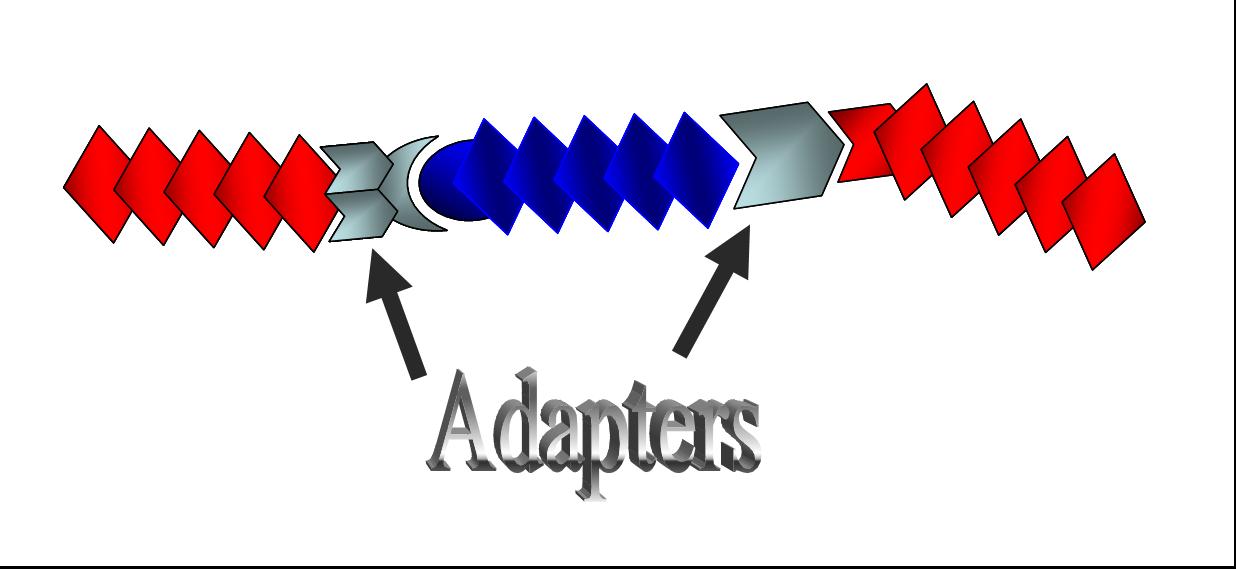
| + | [[Image:adapter2.jpg|thumb|300px|'''Fig. 2 The concept of adapters''' <br>Adapters will provide more convenient Biobrick-parts-construction methods.|right]] | ||
| - | <br>It is necessary for more convenient BioBrick construction | + | <br> |
| + | <br>It is necessary for more convenient BioBrick construction. | ||
<br>(1) to make room for parts insertion between genes after they are connected | <br>(1) to make room for parts insertion between genes after they are connected | ||
<br>(2) to connect genes processed by different restriction enzymes. | <br>(2) to connect genes processed by different restriction enzymes. | ||
| Line 40: | Line 58: | ||
<!--adapaterは完成したのか?完成せずに設計のみならばdevelopよりもdesignの気がする。<実物はないのでしたっけ。--> | <!--adapaterは完成したのか?完成せずに設計のみならばdevelopよりもdesignの気がする。<実物はないのでしたっけ。--> | ||
| - | <br>Together with | + | <br>Together with [http://parts2.mit.edu/wiki/index.php/Tokyo_Alliance:_Design/Method#DNA_Construction_Procedure| Tokyo Standard], the BioBrick parts our last year’s team Tokyo Alliance developed to meet the requirement (1), we suggest a new category of “adapter” as shown in Fig. 2. |
| - | + | ||
| - | + | <br> | |
| - | == | + | ==Future application of our "machines" == |
| - | + | In this iGEM project, we have developed our model using E. coli. | |
| + | The Wet and Dry approaches have demonstrated apparently interesting at the same time academically attracting results. Through these successive Wet and Dry approach and by employing other methods such as a single cell analysis, as mentioned above, we could achieve our model that behaves as we designed. And through this process we could obtain synthetic biologically significant things. | ||
| + | <br><br> | ||
| + | Not only academic issues, could we also apply our "genetically engineered machines" to practical use, or expand our modeled system if we could achieved our goal. | ||
| + | For the practical application, we first focus on the significant feature of the machines - balanced differentiation. | ||
| + | From a single set of cells with the same genetic circuits, we could expect the coexistence of two types of cells and of gene expressions. | ||
| + | <br> | ||
| + | <br> | ||
| + | Consider the case where those two genes encode two different proteins of which concentrations should always remain at a certain ratio. | ||
| + | It is even more convenient if the ratio of the two proteins can be kept only by adding a single set of the machines with the same genes. Regardless of the initial ratio of the two genes being expressed, the machines could change their expressing ways of the genes so that two states of the machines stably coexist. Since the ratio of the machines is kept the same, the ratio of the two expressed proteins would remain the same, though it might show slight oscillation. | ||
| + | <br><br> | ||
| + | In this way, our machine could be applied to biosynthesis of some chemical components and maintenance of the concentrations in solutions at a certain level. | ||
| + | Furthermore, by improving the security and safety, the machines could be medically applied to our human bodies. | ||
| + | There are complex communities bacteria, producing necessary or harmful compounds for us as well as themselves, in our bodies. | ||
| + | Our machines can be applied to some of these intestinal bacteria in order to properly regulate the communities and thereby the production of necessary compounds for us. | ||
| + | <br> | ||
| + | <br> | ||
| + | Not limited to practical applications, we could also improve the machines so that their balanced differentiation is maintained at a much higher or lower concentration, in a larger space, and during longer time course. | ||
| + | Or we could make the machine receive more than two inputs and the whole system stable under the differentiation into three or five different states of the machines. | ||
| + | If we could improve our machines in this way, we could make our model closer to real ecosystems or bacterial communities in the nature. | ||
| + | <br> | ||
| + | <br> | ||
| + | These are merely dreams at this moment; however, the regulation of gene expression depending on the circumstances including other coexisting cells will be better engineered to bring us closer to the dreams. | ||
Latest revision as of 05:26, 27 October 2007
Works top 0.Hybrid promoter 1.Formulation 2.Assay1 3.Simulation 4.Assay2 5.Future works
Balancing gene expression
According to the simulation results and wet experiments, we have found that the relative strength of the promoters of both sides, or relative expression levels of the downstream genes should be changed in order to reach our model.
Even without Dry approaches, it might be possible to get closer to our goal by extremely focusing on such Wet experiments undergoing a number of trial-and-errors.
However, now that we have almost obtained the strong methods of Dry approaches that compensate Wet ones, we can employ even more sophisticated methods as we have done in this project.
We have estimated, based on the simulations and the wet experiments, what should be done next. One option is to change the ribosomal binding sites (RBS) to modulate largely expression level of the downstream genes. There are different RBS available as BioBrick parts, we could choose proper strength of RBS. Another option is that change of -35 and/or -10 sequence of the promoters. Balancing the expression levels of both sides must be a great leap to our goal.
Vice Versa
In the present model, only B de- and redifferentiates in the absence of A but not vice versa.
Establishing complete balanced differentiation model, we have a plan of an improved model where A also dedifferentiates in the absence of B and then redifferentiates into B.
To de- and redifferentiate A, we are planning to use another signaling molecule orthogonal to the AHL for LuxR.
When the ratio of A and B reaches to a certain value, the system is balanced again.
Single cell analysis with a cell sorter
Throughout the Wet experiments so far, we have observed promoter activity of our genetically engineered cells, not at an individual levels but at a collective level. With a single cell analysis, our simulation results would be more clearly confirmed.
We could do this at the next step by using a cell sorter, which separates a mixture of cells into individuals. Then we could observe which states, A, B, or dedifferentiated, individual cells are taking at the moment of the measurement and how many of them are in each state.
Adapters as a new BioBrick category
During the construction of genetic circuits, we often found the necessity of the gene insertion between already ligated genes as in the present work.
However, it is still difficult to exchange iGEM parts, which should be designed convenient for the construction, because iGEM parts have to be ligated laterally.
It is necessary for more convenient BioBrick construction.
(1) to make room for parts insertion between genes after they are connected
(2) to connect genes processed by different restriction enzymes.
Here we suggest “adapters” to solve this inconvenience. In the present project, we have designed “adapters,” BioBrick parts that connect differently digested DNA, meeting the requirement (2).
Together with [http://parts2.mit.edu/wiki/index.php/Tokyo_Alliance:_Design/Method#DNA_Construction_Procedure| Tokyo Standard], the BioBrick parts our last year’s team Tokyo Alliance developed to meet the requirement (1), we suggest a new category of “adapter” as shown in Fig. 2.
Future application of our "machines"
In this iGEM project, we have developed our model using E. coli.
The Wet and Dry approaches have demonstrated apparently interesting at the same time academically attracting results. Through these successive Wet and Dry approach and by employing other methods such as a single cell analysis, as mentioned above, we could achieve our model that behaves as we designed. And through this process we could obtain synthetic biologically significant things.
Not only academic issues, could we also apply our "genetically engineered machines" to practical use, or expand our modeled system if we could achieved our goal.
For the practical application, we first focus on the significant feature of the machines - balanced differentiation.
From a single set of cells with the same genetic circuits, we could expect the coexistence of two types of cells and of gene expressions.
Consider the case where those two genes encode two different proteins of which concentrations should always remain at a certain ratio.
It is even more convenient if the ratio of the two proteins can be kept only by adding a single set of the machines with the same genes. Regardless of the initial ratio of the two genes being expressed, the machines could change their expressing ways of the genes so that two states of the machines stably coexist. Since the ratio of the machines is kept the same, the ratio of the two expressed proteins would remain the same, though it might show slight oscillation.
In this way, our machine could be applied to biosynthesis of some chemical components and maintenance of the concentrations in solutions at a certain level.
Furthermore, by improving the security and safety, the machines could be medically applied to our human bodies.
There are complex communities bacteria, producing necessary or harmful compounds for us as well as themselves, in our bodies.
Our machines can be applied to some of these intestinal bacteria in order to properly regulate the communities and thereby the production of necessary compounds for us.
Not limited to practical applications, we could also improve the machines so that their balanced differentiation is maintained at a much higher or lower concentration, in a larger space, and during longer time course.
Or we could make the machine receive more than two inputs and the whole system stable under the differentiation into three or five different states of the machines.
If we could improve our machines in this way, we could make our model closer to real ecosystems or bacterial communities in the nature.
These are merely dreams at this moment; however, the regulation of gene expression depending on the circumstances including other coexisting cells will be better engineered to bring us closer to the dreams.