From 2007.igem.org
yesterday -- tomorrow
Bastille's Day!!!
What to do ?
- Remove the plate of the transduction experiment and the transformation ones from the 37°C incubator (Upper-left shelf): If it has worked and if someones comes on Sunday:
- isolation of the clones from the transduction on petri-dishes (LB+DAP+Erm+Citrate)
- LB + antibio culture of the transformants (to make a glycerol stock and to do MiniPreps)
- Make a gel (0.8%) and migrate the PCR products
- Purify them
- Do the assembly PCR
If enough time (we can also wait to have the plasmids to do everything at the same time):
- Digestion of the purifications products with appropriate enzymes
- Purification
What has been done
- Several colonies of transduced MG1655 are obtained (transduction from dapA- strain w121):
- Isolation of 10 different colonies on LB Citrate Erythro DAP plates
- Seeding LB-Amp cultures of the following strains:
- DH5alpha transformed with the plasmid carrying the Biobrick pJ23100
- DH5alpha transformed with the plasmid carrying the Biobrick B0015
- These two overnight cultures will allow us to perform (tomorrow):
- Minipreps to isolate the plasmids carrying the biobricks
- Glycerol stocks of the transformed strains
DGAT expressing E.coli
After transformation of DH5alpha E.coli with pKS::DGAT plasmid, a transformant clone was grown overnight in LB Ampicilline medium.
The culture (using a toothpick) was deposited on several solid LB media for growth:
+/- IPTG (inducer of DGAT expression, dgat gene being carried by pKS::DGAT plasmid)
+/- Nile Red (fat detection dye)
+/- oleate at 2 different concentrations (0.5 and 2mM)
PCRs
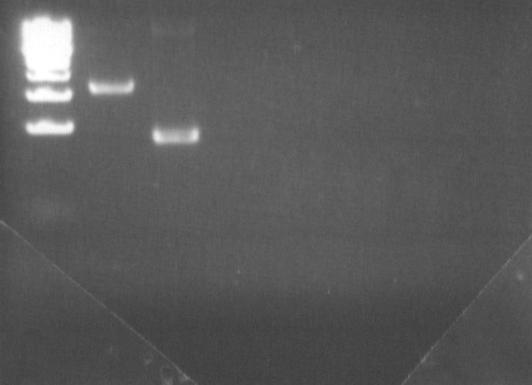
These two first PCRs aims at removing the Pst1 site in DGAT gene.
| PCR : fw-DGAT1 rev-deltaPst1
|
|
|
|
|
|
|
|
|
| PCR Settings
|
| Buffer (5x)
| 5x 10µL
|
| Expected size
|
| Annealing (°C)
|
| MgCl2 10µM
| 10µM 0µL
|
|
|
| 55°C
|
| dNTP 10µM
| 10µM 1µL
|
| Success
|
| Time Elongation
|
| Oligo F 10µM
| 27 forDGAT1
| 1µL
|
| YES
|
| 2m00'
|
| Oligo R 10µM
| 13 DPst1-DGAT-R
| 1µL
|
| Image (click to enlarge)
|
| Number cycles
|
| Water
| 34µL
|
| 
|
| 30
|
| Polymerase
| Phusion 0.5µL
|
| Band (0=ladder)
|
|
|
| DNA
| plasmid pKs::DGAT
|
| 1
|
| PCR : fw_deltaPst1 rev-DGAT1
|
|
|
|
|
|
|
|
|
| PCR Settings
|
| Buffer (5x)
| 5x 10µL
|
| Expected size
|
| Annealing (°C)
|
| MgCl2 10µM
| 10µM 0µL
|
|
|
| 55°C
|
| dNTP 10µM
| 10µM 1µL
|
| Success
|
| Time Elongation
|
| Oligo F 10µM
| 12 fw_deltaPst1
| 1µL
|
| YES
|
| 2m00'
|
| Oligo R 10µM
| 28 RevDGAT1
| 1µL
|
| Image (click to enlarge)
|
| Number cycles
|
| Water
| 34µL
|
| 
|
| 30
|
| Polymerase
| Phusion 0.5µL
|
| Band (0=ladder)
|
|
|
| DNA
| plasmid pKs::DGAT
|
| 2
|
| PCR : Lox71-FtsA-FtsZ-1
|
|
|
|
|
|
|
|
|
| PCR Settings
|
| Buffer (5x)
| 5x 10µL
|
| Expected size
|
| Annealing (°C)
|
| MgCl2 10µM
| 10µM 0µL
|
|
|
| 55°C
|
| dNTP 10µM
| 10µM 1µL
|
| Success
|
| Time Elongation
|
| Oligo F 10µM
| 3 Lox71-FtsA-F
| 2.5µL
|
| YES
|
| 2m00'
|
| Oligo R 10µM
| 4 DEcoR1-FtsZ-R
| 2.5µL
|
| Image (click to enlarge)
|
| Number cycles
|
| Water
| 34µL
|
| 
|
| 30
|
| Polymerase
| Phusion 0.5µL
|
| Band (0=ladder)
|
|
|
| DNA
| toothpick in glycerol stock of MG1655
|
| 1-2
|
| PCR : FtsZ-2
|
|
|
|
|
|
|
|
|
| PCR Settings
|
| Buffer (5x)
| 5x 10µL
|
| Expected size
|
| Annealing (°C)
|
| MgCl2 10µM
| 10µM 0µL
|
|
|
| 55°C
|
| dNTP 10µM
| 10µM 1µL
|
| Success
|
| Time Elongation
|
| Oligo F 10µM
| 5 DEcoR1-FtsZ-F
| 10µM 2.5µL
|
| YES
|
| 2m00'
|
| Oligo R 10µM
| 2 FtsZ-R
| 10µM 2.5µL
|
| Image (click to enlarge)
|
| Number cycles
|
| Water
| 34µL
|
| 
|
| 30
|
| Polymerase
| Phusion 0.5µL
|
| Band (0=ladder)
|
|
|
| DNA
| toothpick in glycerol stock of MG1655
|
| 3-4
|
| PCR : Lox66-DapAColi
|
|
|
|
|
|
|
|
|
| PCR Settings
|
| Buffer (5x)
| 5x 10µL
|
| Expected size
|
| Annealing (°C)
|
| MgCl2 10µM
| 10µM 0µL
|
|
|
| 55°C
|
| dNTP 10µM
| 10µM 1µL
|
| Success
|
| Time Elongation
|
| Oligo F 10µM
| 6 Lox66-DapAColi-F
| 10µM 2.5µL
|
| No
|
| 2m00'
|
| Oligo R 10µM
| 7 DapAColi-R
| 10µM 2.5µL
|
| Image (click to enlarge)
|
| Number cycles
|
| Water
| 34µL
|
| 
|
| 30
|
| Polymerase
| Phusion 0.5µL
|
| Band (0=ladder)
|
|
|
| DNA
| toothpick in glycerol stock of MG1655
|
| 5-6
|


