Paris/July 26
From 2007.igem.org
Colonies PCR for cloning verification
L1.1, L1.2, L2.1, L2.2, L9.1, L10.1
L6: 8 clones
Toothpick in 50µl sterile water ==> 10'@100°C
PCR Mix:
- 212.5 µl Master Mix
- 8.5 µl VF2
- 8.5 µl VR
12 µl of the mix in 13µl of lysed cells for each PCR reaction
Cycle x30:
- 95°C 30
- 60°C 30
- 68°C 2'
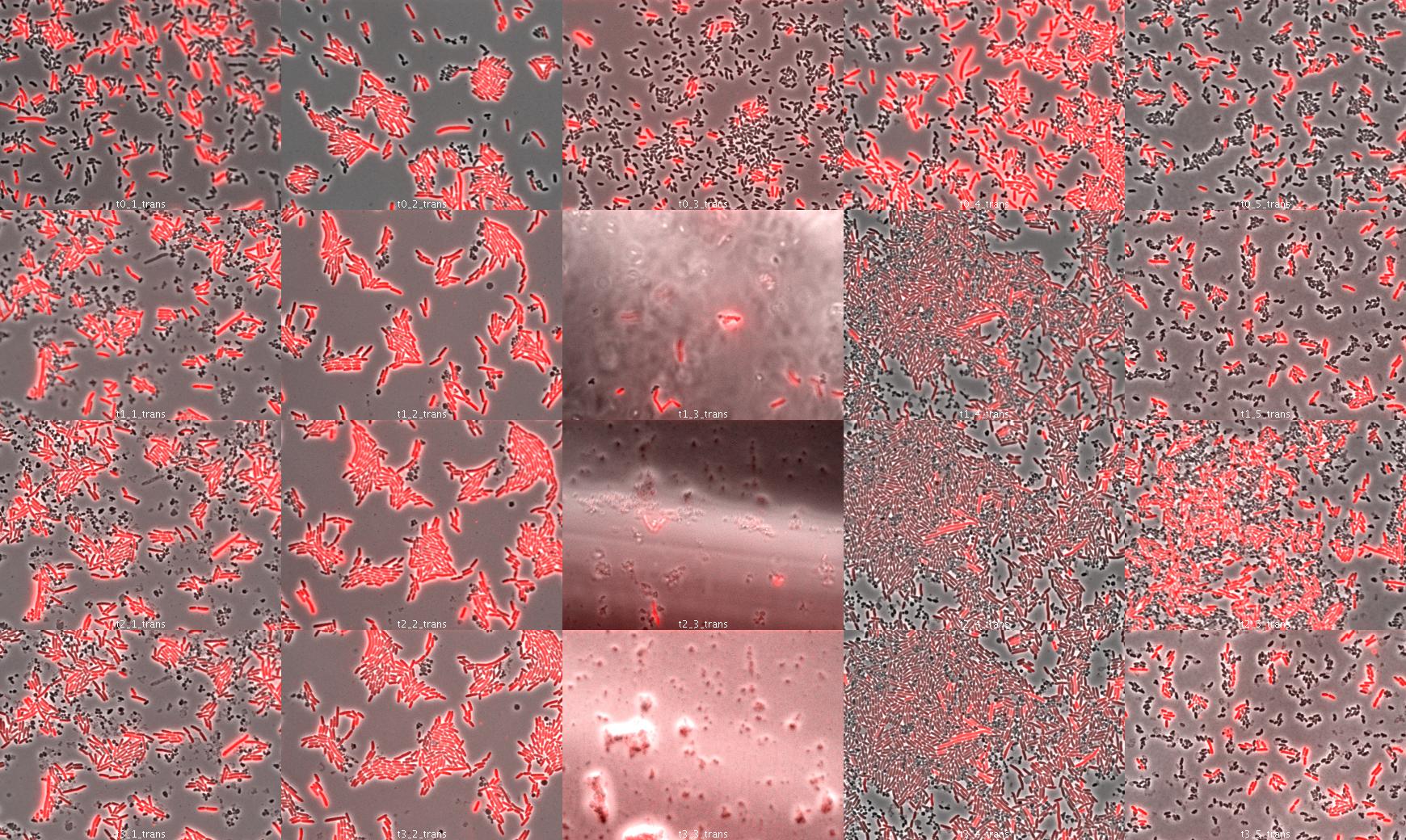
Microscopic DAP complementation experiment
We used wild type E.coli transformed by part J23107 (RFP) and dapA- E.coli (w121) auxotroph to DAP. E.coli transformed by part J23107 (RFP) is able to synthesize DAP and export it. We suppose dap- E.coli could be feed by wt E.coli.
We tried to observe kinetics by fluorescence microscopy. We let separately in culture overnight dapA- E.coli (LB/erythromycine/DAP) and rfp wt E.coli (LB ampicillin). The day after, we diluted both liquid culture 1/200 to have exponential culture.
Then we put on slide different conditions:
- 50% wt rfp / 50% dapA- (colomn 1)
- 70% wt rfp / 30% dapA- (colomn 2)
- 30% wt rfp / 70% dapA- (colomn 3)
- 90% wt rfp / 10% dapA- (colomn 4)
- 10% wt rfp / 90% dapA- (colomn 5)
Under microscope and at 37°C we observed every 30 minutes the bacteria:
The dapA- bacteria do not look to survive; we can observe natural selection directly: rfp wt cells have higher fitness: they multiply
much more than dapA- cells. If dapA- cells were growing, it would have been as parasites because wt cells do not need dapA- cells. So this is just a preliminary experiment because in our system, cells able to multiply need cells unable because of high dapA secretion. This was to see how many wt cells are needed to dapA cells to survive.
We were observing cells on slide under microscope. They do not look to multiply very fast (3-4 divisions in 2 hours). It is possible that dapA cells do not survive because they already are stressed by culture conditions. So we would like to do the same in liquid conditions (we take samples every hour).