Paris/September 3
From 2007.igem.org
dapA deletion in MG1655 K12 E.coli
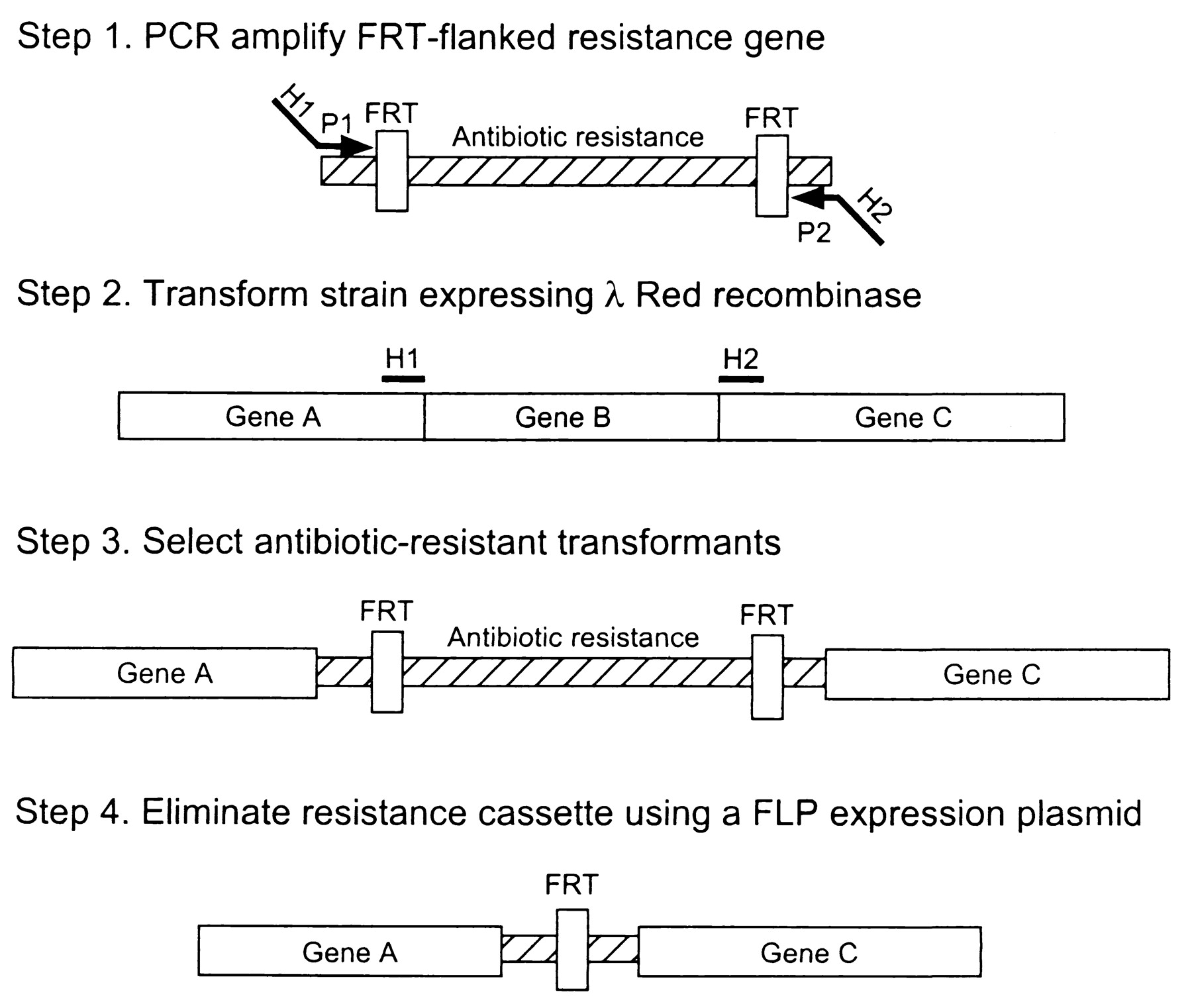
We would like to delete the dapA gene in the chromosome of E.coli K12 (substrain MG1655) with the method of Datsenko and Wanner (One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Datsenko KA, Wanner BL. PNAS 2000: 6640-5, [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=pubmed&dopt=AbstractPlus&list_uids=10829079]) because the transduction method have not worked.
This method is based on homologous recombination using lambda red system. The knocked out gene is replaced with antibiotic resistance gene (kanamycin or chloramphenicol) that is generated by PCR by using primers with 50 nt hommolgy extension. This is accomplished by Red-mediated recombination in this flanking homologies. After selection, the resistance gene can be eliminated by using a helper plasmid expressing the FLP recombinase which acts on the directly repeated FRT sites flanking the resistance gene. The red and FLP helper plasmid can be simply cures by growth at 37°C because they are temperature sensitive replicons.
To do that, we used the protocol from OpenWetWare Recombineering/Lambda red-mediated gene replacement.
See Protocols
We prepared the Transformation Buffer, Arabinose 1M and designed primers. The primers have internal overlap with the resistance marker (20pb) (pKD3: [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=15554329]) and external overlap (50pb) with the target KO gene (dapA: [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=NC_000913.2&from=2596904&to=2597782&strand=2&dopt=fasta]).