Freiburg07/report Ca sensor
From 2007.igem.org
| Line 82: | Line 82: | ||
pr-calmo-SpeI-linker6: 5' AACGATCACT AGTACCGCCA CCAGAGCCAC CCTTTGCTGT CATCATTTGT ACA 3'<br> | pr-calmo-SpeI-linker6: 5' AACGATCACT AGTACCGCCA CCAGAGCCAC CCTTTGCTGT CATCATTTGT ACA 3'<br> | ||
pr-calmo-SpeI-linker9: 5' AACGATCACT AGTACCGCCA CCGCCAGAAC CGCCACCGCC CTTTGCTGTC ATCATTTGTA CA 3'<br> | pr-calmo-SpeI-linker9: 5' AACGATCACT AGTACCGCCA CCGCCAGAAC CGCCACCGCC CTTTGCTGTC ATCATTTGTA CA 3'<br> | ||
| + | |||
| + | |||
| + | |||
<div style="float:right"> | <div style="float:right"> | ||
Revision as of 19:21, 25 October 2007
Contents |
A single-protein calcium ion sensor mediating cell survival
Introduction
The idea:
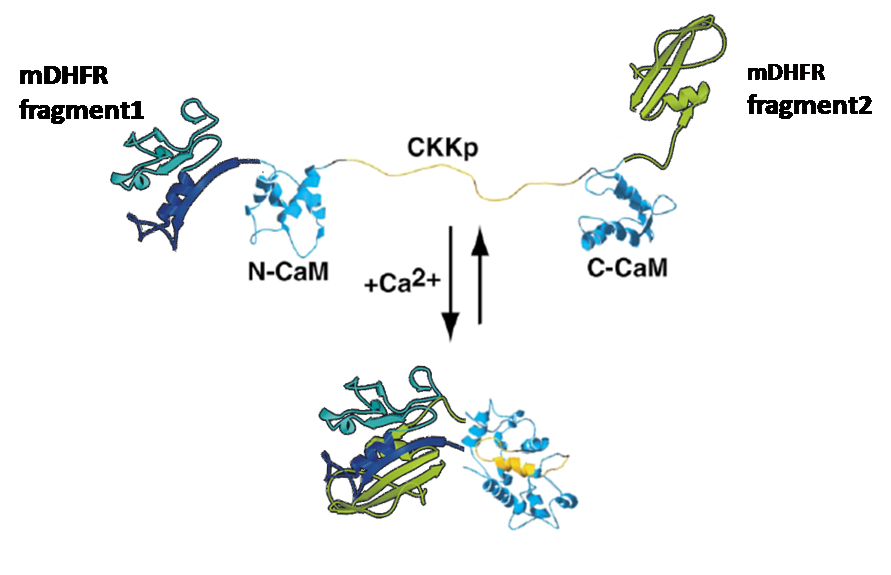
Was to attach enzyme-halfs to several trigger-proteins (here: calmodulin)
by cloning each one of the enzyme-halfs and the trigger-protein together in one plasmid.
This plasmid would then contain the code for the whole "protein-machinery", allowing in vivo-tests as well as expression and purification of the engineered protein, an on/off-switchable enzyme.
Split enzymes:
Some enzymes can be divided into two separate parts that don´t show any activity until both parts are physically brought together;
this circumstance allows enzyme-activity-assays.
As those assays were already being used in our lab before, we could access plasmids containing enzyme-halfs of beta-lactamase and dihydrofolatereductase (DHFR).
Calmodulin:
This molecule is known for its strong conformational change upon binding calcium.
Truong, Ikura et al.(Ontario Cancer Institute and [http://medbio.utoronto.ca/ Department of medical Biophysics] , University of Toronto) have fused CFP and YFP to the ends of modified calmodulin and shown that you can then induce [http://en.wikipedia.org/wiki/Fluorescence_resonance_energy_transfer FRET] between them by adding calcium.
This work inspired us to test calmodulin as "switch" for our split enzymes. Prof. Ikura/ Le Zheng were so friendly to send us this calmodulin (YC6.1 in plasmid pcDNA3 with sequence, thanks a lot again!) so that we could gain it via PCR.
Materials and Methods
Planning:
-Plasmid maps, sequences, alignments and protein dates have been worked out using "gcg", a unix-based, rather antiquate genetic engineering program
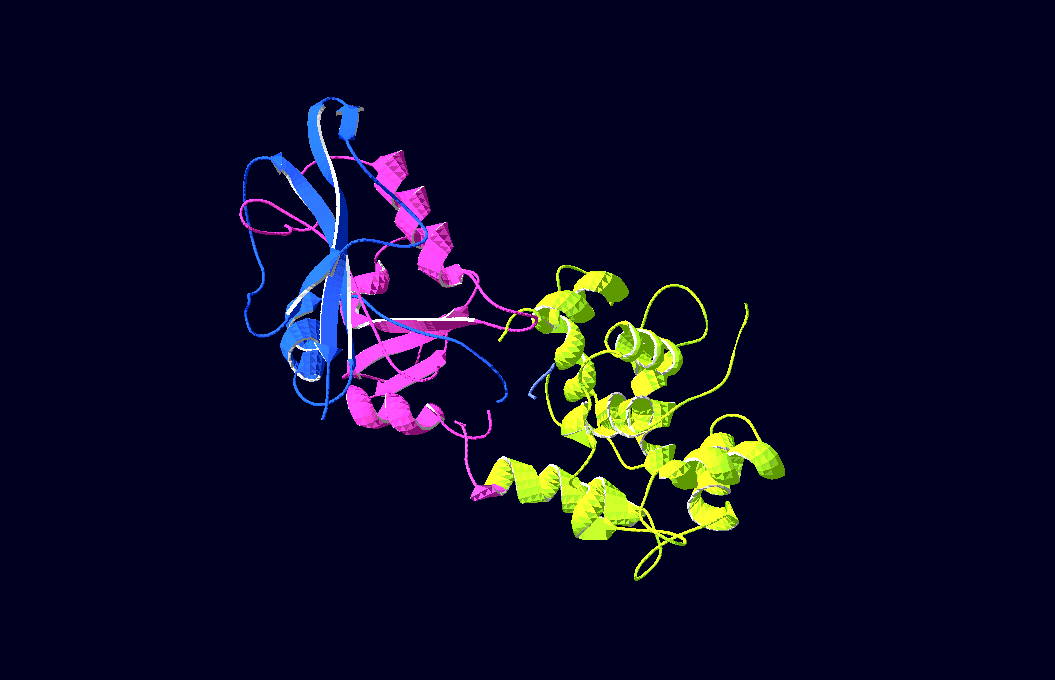
-3D-Models of our constructs (as shown in the pictures above) have been created/modified using (Mac-)Pymol and SwissPdbViewer (both freeware:[http://pymol.sourceforge.net/ Pymol],[http://expasy.org/spdbv/ SwissPdbViewer])
Cloning:
-Cloning steps have been done as described under protocols or with Qiagen-products
Protein expression and purification:
-Expression cultures (Volume: 2 liters) have been raised at 37 deg. C and induced by IPTG
-Protein purifications have been carried out via Ni-NTA-columns; the His-Tagged proteins have been eluted in buffers with rising imidazole-concentrations and were then analyzed on SDS-gels
Mediums and Plates
DNA sequencing
Ligation
Plasmid spin column prep
Glycerol stocks
General Gene-Protein Information
Purification
Transformation
Dephosphorylation
Preparative Digestion
Analytic Digestion
Gel Electrophoresis
Polyacrylamide gel electrophoresis
Protein purification
In vivo test I
In vivo test II
Calmodulin via PCR:
forward primers:
pf-calmo-KpnI-linker2: 5' TATCGACGGT ACCGGCGGTG AGCAGATTGC AGAGTTCAAA G 3'
pf-calmo-KpnI-linker3: 5' TATCGACGGT ACCGGCGGTG GCGAGCAGAT TGCAGAGTTC AAAG 3'
pf-calmo-KpnI-linker4: 5' TATCGACGGT ACCGGCGGTG GCGGTGAGCA GATTGCAGAG TTCAAAG 3'
pf-calmo-KpnI-linker6: 5' TATCGACGGT ACCGGCGGTG GCTCTGGTGG CGAGCAGATT GCAGAGTTCA AAG 3'
pf-calmo-KpnI-linker9: 5' TATCGACGGT ACCGGCGGTG GCGGTTCTGG CGGTGGCGGT GAGCAGATTG CAGAGTTCAA AG 3'
reverse primers:
pr-calmo-SpeI-linker2: 5' AACGATCACT AGTACCTCCC TTTGCTGTCA TCATTTGTAC A 3'
pr-calmo-SpeI-linker3: 5' AACGATCACT AGTACCGCCA CCGCCCTTTG CTGTCATCAT TTGTACA 3'
pr-calmo-SpeI-linker4: 5' AACGATCACT AGTGCCACCG CCTCCCTTTG CTGTCATCAT TTGTACA 3'
pr-calmo-SpeI-linker6: 5' AACGATCACT AGTACCGCCA CCAGAGCCAC CCTTTGCTGT CATCATTTGT ACA 3'
pr-calmo-SpeI-linker9: 5' AACGATCACT AGTACCGCCA CCGCCAGAAC CGCCACCGCC CTTTGCTGTC ATCATTTGTA CA 3'
Results
Plasmids:
Ca2+ sensor: ß-lactamase
Plasmids with calmodulin between the lactamase fragments:
2 amino acids for linker: pFR320p_b1_2calmo2_b2
4 amino acids for linker: pFR320p_b1_4calmo4_b2
6 amino acids for linker: pFR320p_b1_6calmo6_b2
Ca2+ sensor: DHFR
Plasmids with calmodulin between the dhfr fragments:
2 amino acids for linker: dFR320d-dhfr1-2calmo2-dhfr2A
4 amino acids for linker: dFR320d-dhfr1-4calmo4-dhfr2A
6 amino acids for linker: dFR320d-dhfr1-6calmo6-dhfr2A
We gained plasmids containing both parts of each split-enzyme, linked to calmoduline by 2/4/6 glycins each;
in-vivo-growth tests showed the best activity for 4-glycine linkers;
thus we decided to proceed
with this linker-length.
In-Vivo-Analysis:
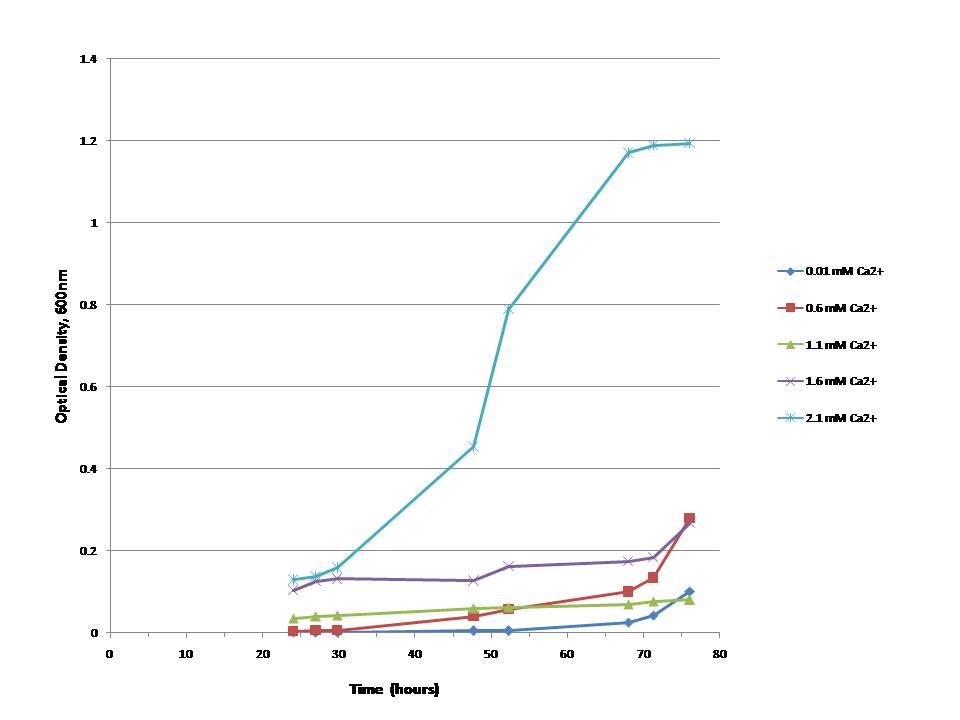
As DHFR is essential for bacterial growth in minimal medium with trimethioprim (TMP),
we could show that the ability of E.colis with our plasmid to grow is directly dependant on the presence -and, furthermore, concentration- of calcium-ions (Ca2+)in the medium:In vivo test II:
In this test we inoculated M9 minimal medium at different calcium concentrations with DHFR1-4Calmodulin4-DHFR2A-positive E. colis and measured the absorptions (first measurement after 24 hours and then again every three to 10 hours for another two days/ 50 hours). Several tests we performed before made this approach seem best.
Discussion
Our goal to create a fusion-protein sensor with functional nano-mechanical aspects seems to be achieved at least for DHFR.
Cloning the beta-lactamase plasmid as planned, we encountered various problems such as unexpected mutations and the circumstance that the product seemed to be toxic for E. coli. For this reason we designed optimized split-lactamase parts to enhance stability.
Parts submitted
References
split DHFR:
Pelletier JN, Arndt KM, Plückthun A, Michnick SW. "An in vivo library-versus-library selection of optimized protein-protein interactions." Nat Biotechnol. 1999 Jul;17(7):683-90.
Pelletier JN, Campbell-Valois FX, Michnick SW. "Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments." Proc Natl Acad Sci U S A. 1998 Oct 13;95(21):12141-6. [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=9770453&ordinalpos=5&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum pubmed]
Calmodulin Sensor:
Pham E, Chiang J, Li I, Shum W, Truong K "A computational tool for designing FRET protein biosensors by rigid-body sampling of their conformational space." Structure. 2007 May;15(5):515-23.
Truong K, Sawano A, Miyawaki A, Ikura M. "Calcium indicators based on calmodulin-fluorescent protein fusions." Methods Mol Biol. 2007;352:71-82.
Truong K, Sawano A, Mizuno H, Hama H, Tong KI, Mal TK, Miyawaki A, Ikura M. "FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule." Nat Struct Biol. 2001 Dec;8(12):1069-73.
Reviews on calmodulin: Grabarek Z "Structural basis for diversity of the EF-hand calcium-binding proteins." J Mol Biol. 2006 Jun 9;359(3):509-25.
Soderling TR, Stull JT "Structure and regulation of calcium/calmodulin-dependent protein kinases." Chem Rev. 2001 Aug;101(8):2341-52