Edinburgh/Yoghurt/Proof of concept
From 2007.igem.org
| (2 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
---- | ---- | ||
[[Image:Edinburgh Uni Logo.jpg|50 px]] | [[Image:Edinburgh Uni Logo.jpg|50 px]] | ||
| - | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/Proof of concept|Proof of concept]] | [[Edinburgh/Yoghurt/References|References]] | + | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/Proof of concept|Proof of concept]] | [[Edinburgh/Future| Future Directions]] | [[Edinburgh/Yoghurt/References|References]] |
---- | ---- | ||
__TOC__ | __TOC__ | ||
| - | In order to test the feasibility of gene expression in ''Lactobacillus'' and the possibility of making self flavouring yoghurt, we required a proof of concept. | + | In order to test the feasibility of gene expression in ''Lactobacillus'' and the possibility of making self-flavouring yoghurt, we required a proof of concept. |
As both the pigment and flavour pathways are rather complex and will require modification and optimisation before their expression in yoghurt we planned to use the much simpler Red Fluorescent Protein (RFP) gene as a proof of concept. | As both the pigment and flavour pathways are rather complex and will require modification and optimisation before their expression in yoghurt we planned to use the much simpler Red Fluorescent Protein (RFP) gene as a proof of concept. | ||
| Line 26: | Line 26: | ||
* this vector was then transformed into ''E. coli'' | * this vector was then transformed into ''E. coli'' | ||
| - | * | + | * red colonies were produced on standard growth media. These can be viewed in Figure 1. |
| Line 36: | Line 36: | ||
RFP synthesis was observed. | RFP synthesis was observed. | ||
| - | Colonies can be viewed in | + | Colonies can be viewed in Figure 2. |
| Line 92: | Line 92: | ||
---- | ---- | ||
| - | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/Proof of concept|Proof of concept]] | [[Edinburgh/Yoghurt/References|References]] | + | [[Edinburgh/Yoghurt| Introduction]] | [[Edinburgh/Yoghurt/Applications|Applications]] | [[Edinburgh/Yoghurt/Design|Design]] | [[Edinburgh/Yoghurt/Modelling|Modelling]] | [[Edinburgh/Yoghurt/Wet Lab|Wet Lab]] | [[Edinburgh/Yoghurt/Proof of concept|Proof of concept]] | [[Edinburgh/Future| Future Directions]] | [[Edinburgh/Yoghurt/References|References]] |
---- | ---- | ||
Latest revision as of 20:26, 26 October 2007
 Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | Future Directions | References
Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | Future Directions | References
Contents |
In order to test the feasibility of gene expression in Lactobacillus and the possibility of making self-flavouring yoghurt, we required a proof of concept.
As both the pigment and flavour pathways are rather complex and will require modification and optimisation before their expression in yoghurt we planned to use the much simpler Red Fluorescent Protein (RFP) gene as a proof of concept.
So far we have managed to transform a number of RFP-pTG262 vectors containing a variety of promoters into E. coli and also Bacillus subtilis, a gram positive bacterium.
We chose to initially transform Bacillus subtilis with our proof of concept vectors over Lactobacillus for two reasons:
- 1. the bacterium is naturally competent at certain stages of its life cycle and therefore has a much simpler transformation process.
- 2. members of the French lab have previous experience of transforming Bacillus subtilis.
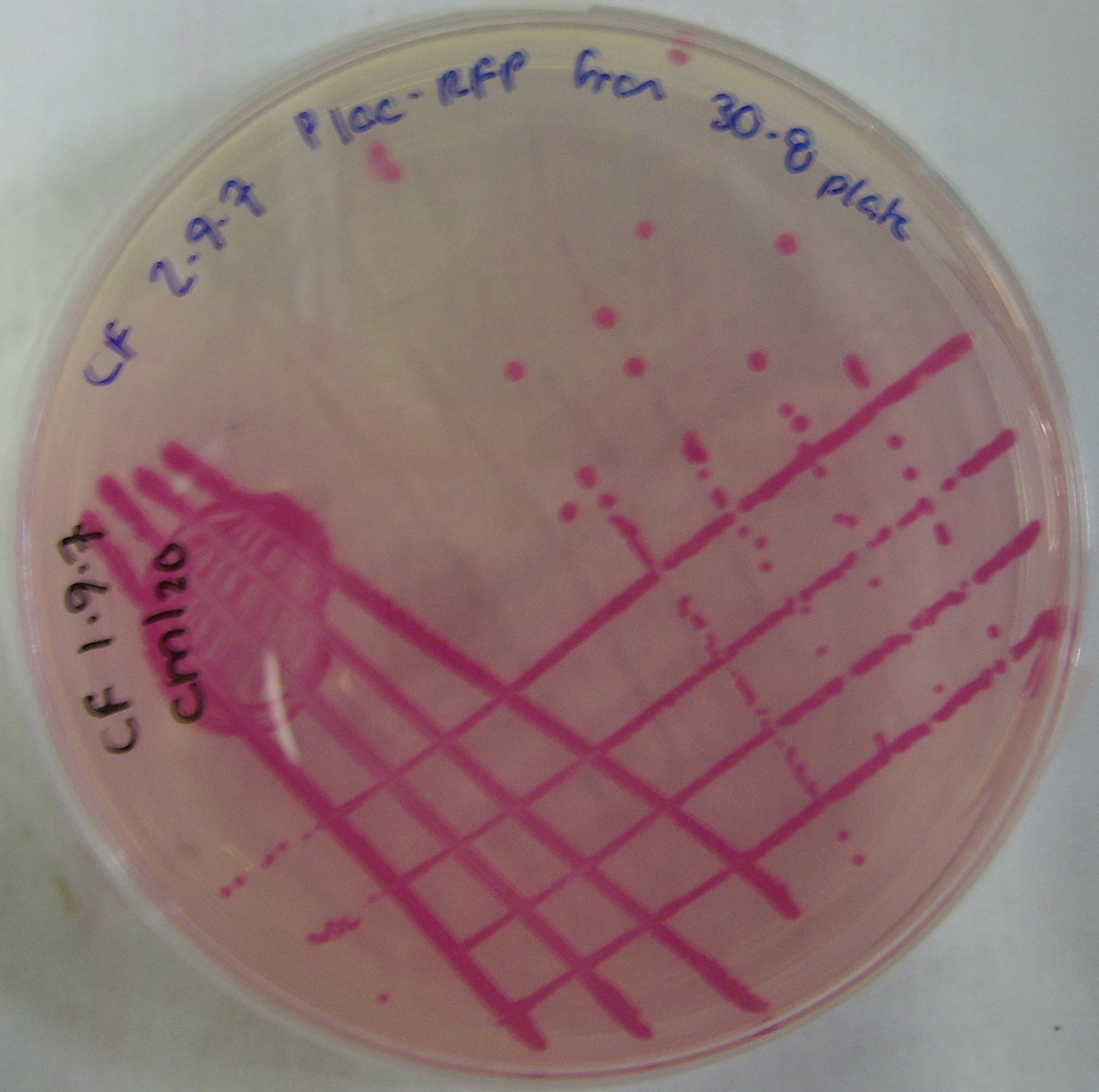
pTG262-PLac-RFP construct
We have inserted the lactose induced RFP gene into the pTG262 vector
- this vector was then transformed into E. coli
- red colonies were produced on standard growth media. These can be viewed in Figure 1.
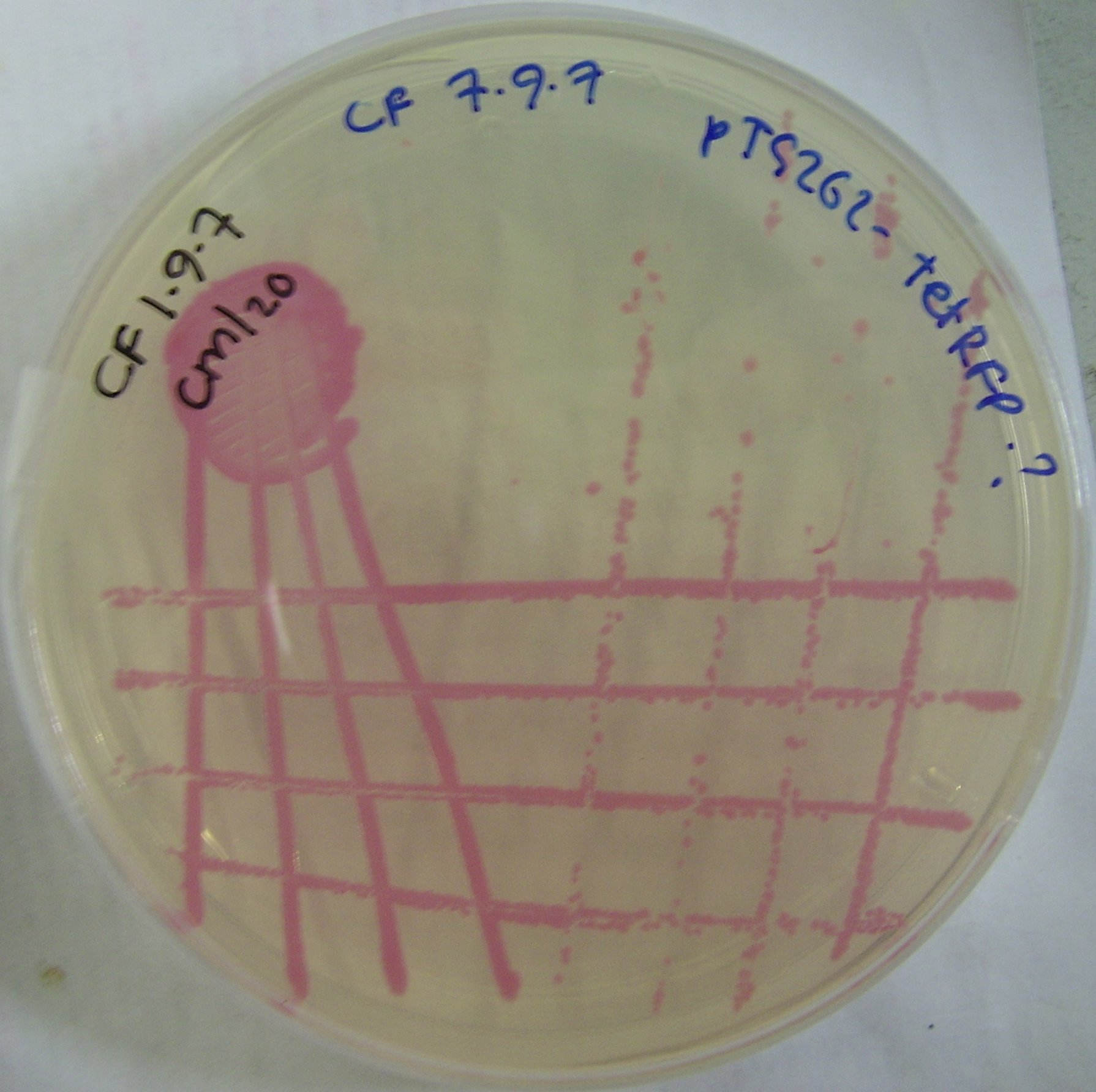
pTG262-Ptet-RFP construct
We ligated the Ptet induced RFP biobrick into the pTG262 vector. We then transformed the vector into E. coli.
RFP synthesis was observed.
Colonies can be viewed in Figure 2.
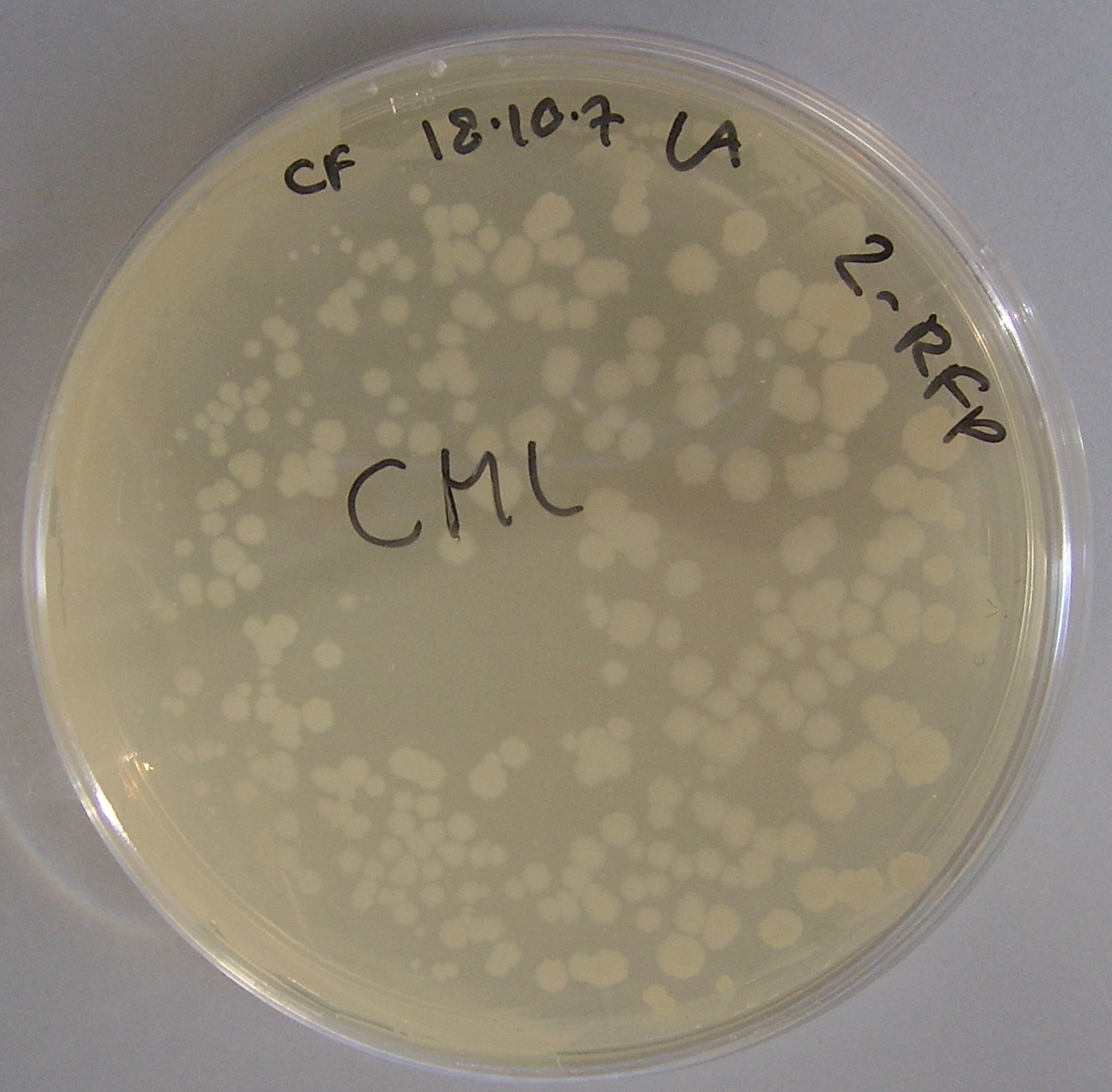
pTG262 expression in Bacillus subtilis
After the successful expression of both the pTG262 RFP constructs in E. coli, we decided to determine if pTG262 could be stably maintained in Bacillus subtilis.
To do this we transformed both the pTG262 and Plac-RFP-pTG262 vectors into Bacillus subtilis.
Figure 3 depicts the successful transformation and expression of pTG262 in Bacillus subtilis.
This is determined by the ability of Bacillus to grow on chloramphenicol plates. No growth was observed on negative control plates.
Unfortunately RFP was not expressed from the promoter, as shown by the colonies being white rather than red. We are attributing this to the Bacillus transcription machinery (particularly RNA polymerase and its associated sigma factors) not recognising the promoter.
Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | Future Directions | References