Edinburgh/Yoghurt/Design
From 2007.igem.org
 Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | References
Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | References
Contents |
Colour Production
The zeaxanthin operon is naturally found in many plants and bacteria. The operon includes five genes, which enable the production of the yellow pigment zeaxantin.
The five genes present in the zeaxanthin operon are:
- crtE
- crtB
- crtI
- crtY
- crtZ
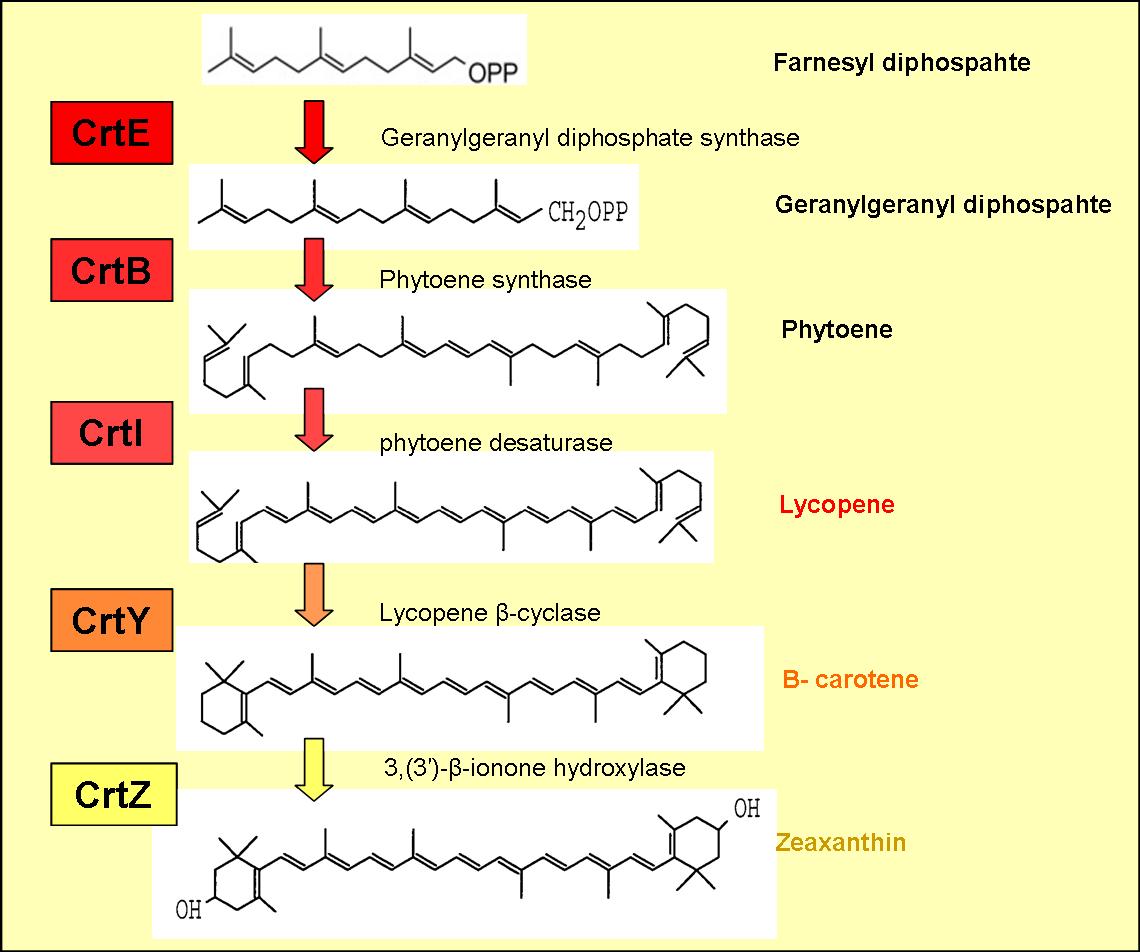
The enzymes encoded by these genes and the products formed are displayed in figure 1.
There are three pigments produced by the genes with in the zeaxanthin operon and a brief description of each is given below:
Lycopene
The enzymes CrtE, CrtB and CrtI produce lycopene from farnesyl diphosphate (an intermediate in the mevalonate pathway).
Lycopene is a red pigment (found in tomatoes), which also has extremely powerful antioxidant properties and may possibly help protect against cancer.
Biobrick may be found under [http://partsregistry.org/Part:BBa_I742120 BBa_I742120] and [http://partsregistry.org/Part:BBa_I742136 BBa_I742136]
Beta-carotene
Beta-carotene is produced by cyclising lycopene, which is carried out by lycopene B-cyclase encoded by the gene crtY (see figure )
B-carotene has an orange pigmentation and is responsible for the colour of carrots, winter squash and several other vegetables. The pigment can be stored in the liver and converted to Vitamin A, a form of retinol, required for sight.
Biobricks may be found under [http://partsregistry.org/Part:BBa_I742138 BBa_I742138] and [http://partsregistry.org/Part:BBa_I742121 BBa_I742121] in the registry
Zeaxanthin
Addition of crtZ to the crtEBIY construct enables the hydroxylation of Beta-carotene (see figure 1) to zeaxanthin, which is a yellow pigment.
Proposed Biobricks
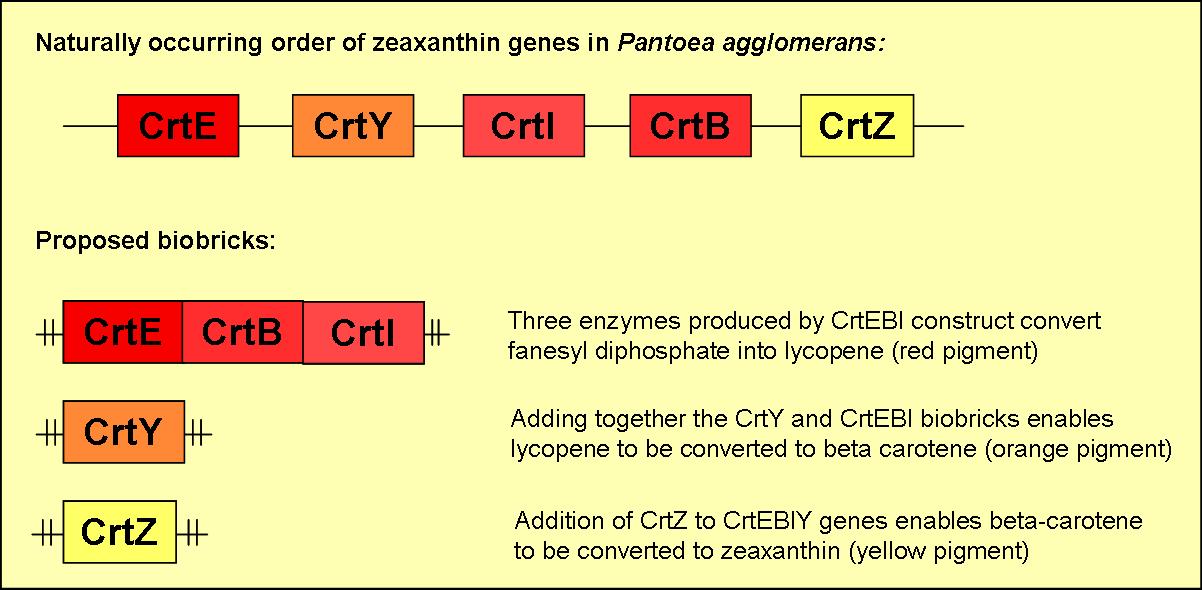
We plan to produce five different biobricks with varying combinations of the five zeaxanthin genes. A brief overview of three of these biobricks can be found in figure 2.
Biobrick 1
- will consist of the three genes crtE, crtB & crtI
- will produce the red pigment lycopene
- [http://partsregistry.org/Part:BBa_I742120 BBa_I742120] and [http://partsregistry.org/Part:BBa_I742136 BBa_I742136]
Biobrick 2
- will contain crtY
- when induced in the presence of functional crtEBI genes an orange pigment (beta-carotene) will be formed
- [http://partsregistry.org/Part:BBa_I742117 BBa_I742117]
Biobrick 3
- will consist of crtZ
- when induced in the presence of functional crtEBIY genes it will enable the formation of the yellow pigment zeaxanthin
- [http://partsregistry.org/Part:BBa_I742118 BBa_I742118]
Biobrick 4
- will contain the four genes crtE, crtB, crtI & crtY
- the construct will be capable of producing beta-carotene
Biobrick 5
- will contain all five genes present in the zeaxanthin operon; crtE, crtB, crtI, crtY & crtZ
- biobrick will enable the production of zeaxanthin (yellow pigmentation)
- [http://partsregistry.org/Part:BBa_I742122 BBa_I742122] and [http://partsregistry.org/Part:BBa_I742139 BBa_I742139]
Vanilla Flavour Production
In order to create vanilla flavouring for our yoghurt, we have designed a novel vanillin biosynthesis pathway. The pathway we devised consists of five different genes, which enable the synthesis of vanillin from the amino acid tyrosine.
We chose to use tyrosine as a starting point, as the amino acid is produced endogenously by bacteria, and is also present within milk proteins. Theoretically this means a starting substrate will not have to be added to the yoghurt starter culture, to faciliate the synthesis of vanillin.
The vanillin biosynthesis pathway was created by piecing together five different genes from three completely different organisms. These five genes are:
- sam8
- sam5
- COMT
- fcs
- ech
Final construct of all five genes may be viewed in the registry under [http://partsregistry.org/Part:BBa_I742140 BBa_I742140]
sam8
- isolated from the bacterium Saccharothrix espeanensis
- encodes tyrosine ammonia lyase (catalyses the deamination of tyrosine's amine group)
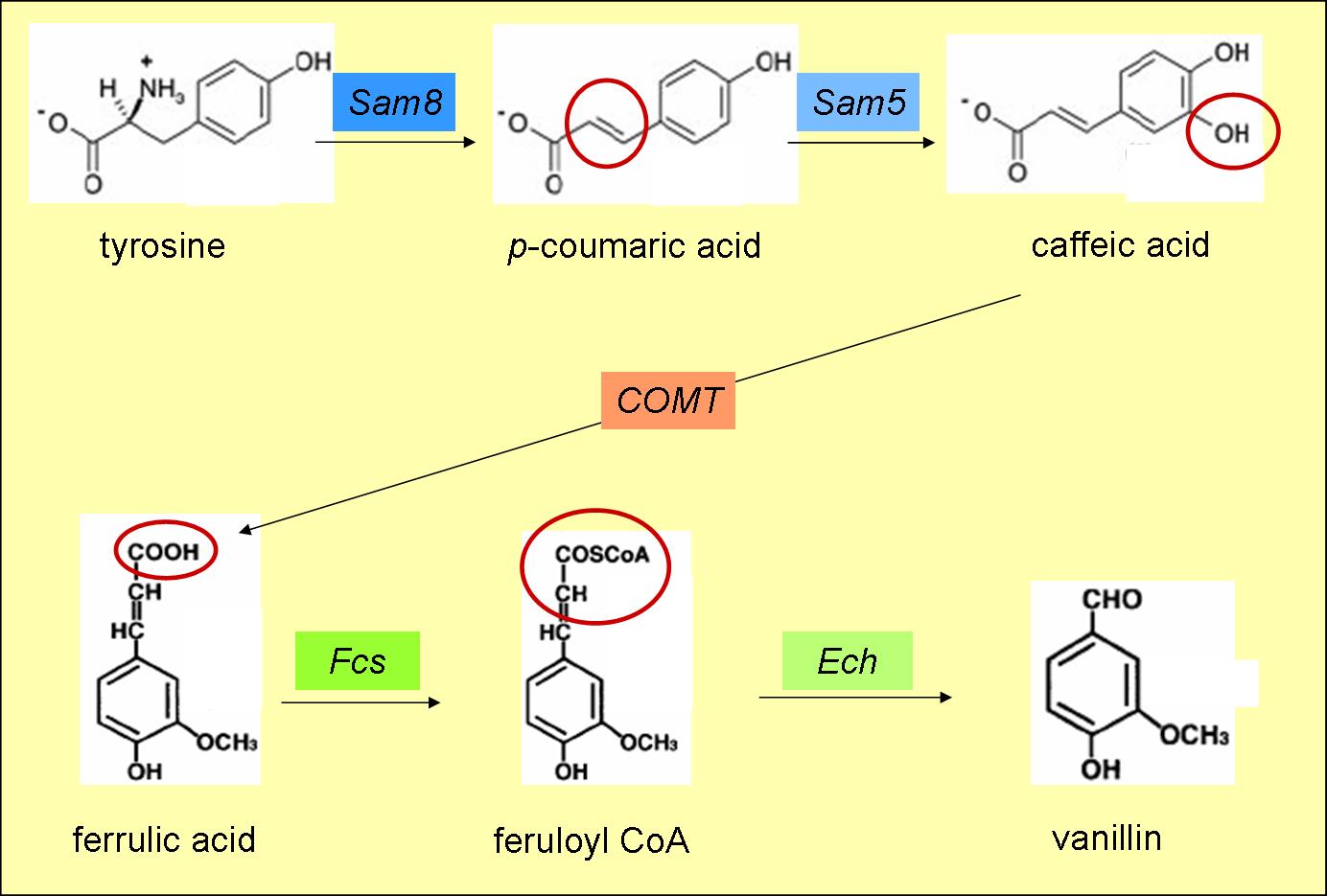
- converts tyrosine to p-coumaric acid (fig. 3)
- catalyses the first step in the vanillin biosynthesis pathway
- sam8 part can be found in the registry under [http://partsregistry.org/Part:BBa_I742142 BBa_I742142]
sam5
- also isolated from Saccharothrix espeanensis
- encodes 4-coumarate 3-hydroxylase (hydroxylates C4 in the aromatic ring of p-coumaric acid)
- converts p-coumaric acid to caffeic acid (fig. 3)
- catalyses the second step in the vanillin biosynthesis pathway
- Sam5 biobrick may be found in the registry under [http://partsregistry.org/Part:BBa_I742141 BBa_I742141]
COMT
- enzyme is natively found in the plant alfalfa
- encodes caffeic acid-O-methyl transferase (methylates -OH on C4 of the aromatic ring)
- produces ferulic acid from caffeic acid (fig. 3)
- catalyses the third step of the vanillin biosynthesis pathway
- COMT biobrick may be found in the registry under [http://partsregistry.org/Part:BBa_I742109 BBa_I742109]
fcs
- gene was isolated from Pseudomonas fluorescens
- encodes feruoyl CoA synthase (ligates acetyl-CoA onto ferulic acid)
- produces feruloyl CoA from ferrulic acid (fig. 3)
- catalyses the penultimate of the vanillin biosynthesis pathway
- Fcs biobrick may be found in the registry under [http://partsregistry.org/Part:BBa_I742113 BBa_I742113]
ech
- gene was also isolated from Pseudomonas fluorescens
- encodes enoyl CoA hydratase (cleaves CoA group from feruloyl CoA)
- converts feruloyl CoA to vanillin (fig. 3)
- catalyses the final stage of the vanillin biosynthesis pathway
- Ech biobrick may be found in the registry under [http://partsregistry.org/Part:BBa_I742115 BBa_I742115]
Proposed Biobricks
We plan to construct the vanillin pathway in three separate constructs:
Construct 1
- consist of sam5 and sam8 genes ligated together with ribosome binding sites
- we will test the construct for the production of caffeic acid from tyrosine
Construct 2
- will only contain the COMT gene ordered from GENEART, together with ribosome binding site
- we will test the production of ferulic acid from caffeic acid
Construct 3
- will contain the ech and fcs genes along with ribosome binding sites
- two genes will be tested for the formation of vanillin from ferulic acid
Lemon Flavour Production
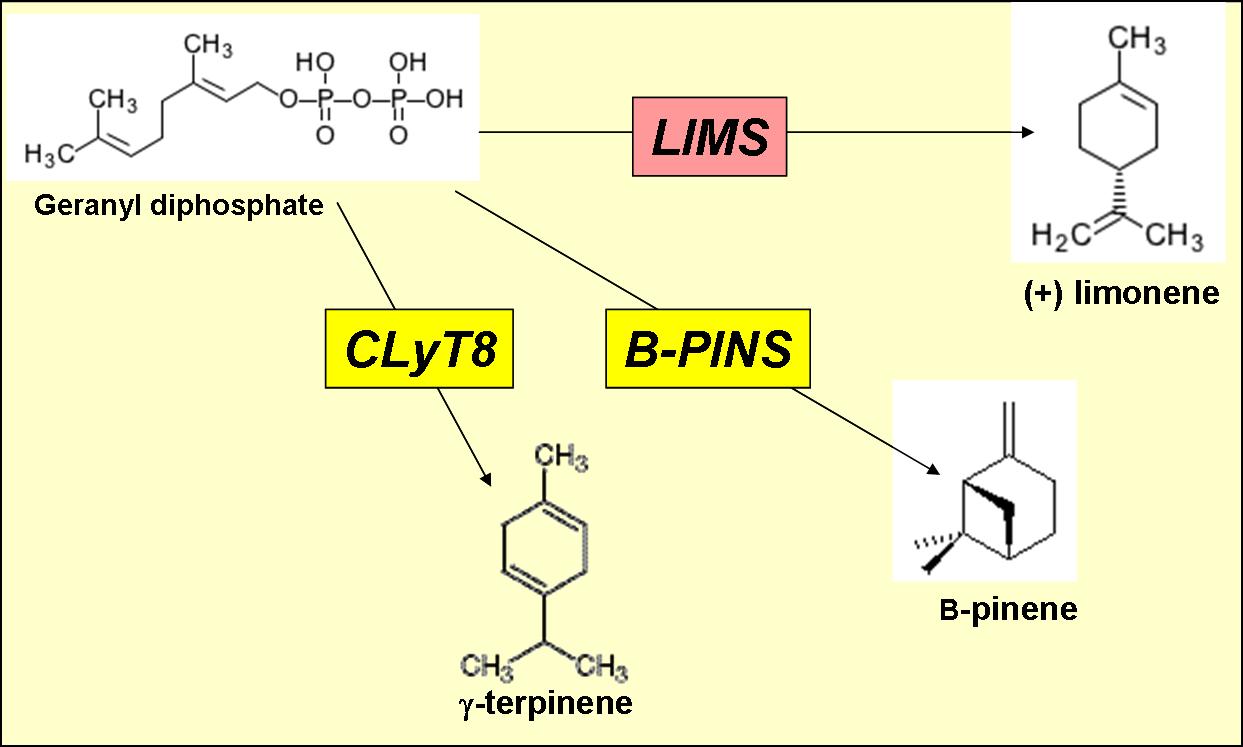
The second flavour we thought of synthetically synthesing is lemon. Lemons produce a wide variety and high quantity of monoterpenoids from the precursor geranyl diphosphate.
Some reasons for choosing lemon:
- several lemon monoterpenoid genes responsible for flavour have recently been identified & characterised
- lemon oil is the highest value essential oil annually imported into the United States
- lemon flavour is used in a wide variety of applications, such as fragrance, flavouring and pharmaceuticals
Several studies of lemon flavour and aroma indicate that three monoterpenoids contribute towards the majority of lemon flavour.
- (+) limonene (synthesised by limonene synthase)
- gamma-terpinene (synthesised by gamma-terpinene synthase)
- (-) beta-pinene (synthesised by beta-pinene synthase)
The flavour compounds produced by lemons are sterospecific, highlighting the importance of their synthetic production by enzymes, which carry out sterospecific reactions, rather than chemical synthesis.
Unfortunately all three of the genes responsible for lemon flavour synthesis have three or more forbidden restriction sites, making their isolation and insertion into biobricks impossible within a short period of time, especially when the issue of potential introns is also considered. Instead we have sent the (+) limonene synthase gene (LIMS1) to GENEART for synthesis. We chose to artifically synthesise the LIMS enzyme as (+) limonene is thought to contribute to ~90% of lemon flavouring. The coding sequence was modified to remove the forbidden restriction sites and to remove the N-terminal plastid-targeting sequence.
The limonene synthase gene may be found in the registry under [http://partsregistry.org/Part:BBa_I742111 BBa_I742111]
Gene Expression In Lactobacillus
In order to express our flavour and colour genes in lactic acid bactia (LAB), we require a different vector to those currently in use by the registry. After some research, we found that a few groups within the UK were working with LAB.
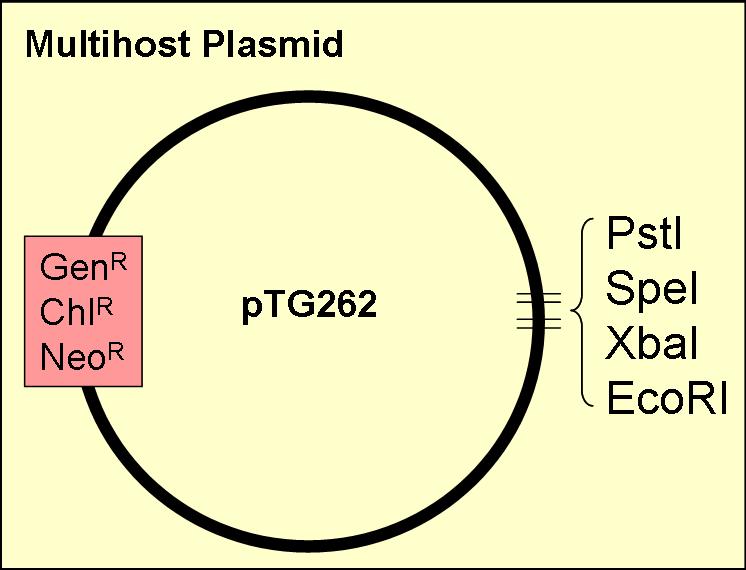
We kindly thank Dr Mike Gasson & Dr. Claire Shearman, of the Institute of Food Research in Norwich for generously donating the pTG262 vector.
pTG262 plasmid is deposited in the registry under [http://partsregistry.org/Part:BBa_I742103 BBa_I742103] and [http://partsregistry.org/Part:BBa_I742123 BBa_I742123]
pTG262
This vector was ideal for use as a biobrick vector, as:
- it contained three of the four biobrick restriction enzyme sites (insertion of a biobrick added the other two)
- it has a gram positive origin of replication, which also works well in E. coli (enabling the plasmid to act as a shuttle vector between E. coli and LAB
- contains three antibiotic resistance genes (chloramphenicol, neomycin & gentamicin)
- known to work in Lactobacillus, Lactococcus and Bacillus as well as E. coli
We also plan to test the efficieny of the vector in other gram negative bacteria including; Shewanella & Pseudomonas, Agrobacterium.
Inducible Gene Expression
In order to have 'self flavouring yoghurt' we needed to find a method of inducing the expression of specific flavour and colour biosynthesis pathways. One method of doing this in food grade products is to use sugar inducible promoters.
During our research we identified three promoters that could possibly induce gene expression, when activated by the presence of a certain sugar. These three promoters are, arabinose, trehalose and maltose. Unfortunately we wouldn't be able to use the lac operon promoter in yoghurt, as lactose is freely available, resulting in continual activation of that particular promoter.
Maltose promoter
Regulated by MalT protein – a positive regulator of all mal genes
MalT is activated by binding ATP and maltotriose.
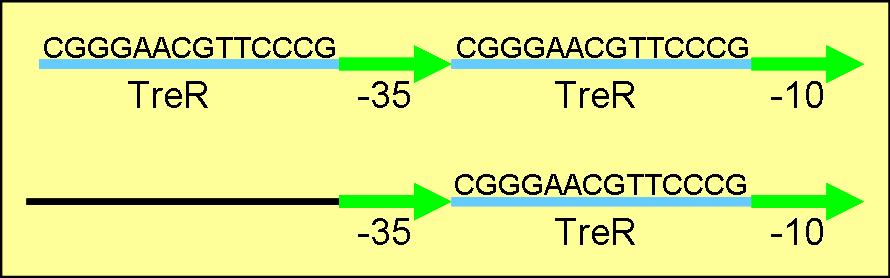
Trehalose promoter
E. Coli trehalose operon is both repressed by TreR protein and under CAP mediated activation
LacI Lambda pL hybrid in registry does not require CAP binding to induce gene expression
Propose to create similar promoter that is repressed in absence of trehalose and does not require CAP to induce transcription.
Replaced lac01 LacI binding site with TreR binding site. Transcription should be induced by the binding of trehalose-6-P to repressor and dissociation of the TreR repressor
Arabinose Promoter
May be found in registry under [http://partsregistry.org/Part:BBa_I0500 BBa_I0500]
Introduction | Applications | Design | Modelling | Wet Lab | Proof of concept | References