Melbourne/Lab Notebook gv 3
From 2007.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | [[Melbourne/Lab GV Notebook| <Return to lab book summary>]] | ||
==Prepared for site dirrected mutagenesis== | ==Prepared for site dirrected mutagenesis== | ||
| - | + | *Produced NZY broth 100mL autoclaved (due to contamination in last batch). | |
| - | + | * Diluted DNA from last round | |
| - | *Produced NZY broth 100mL autoclaved | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | * | + | |
| - | + | ||
{| border="1" | {| border="1" | ||
| - | |||
|- | |- | ||
| - | ! | + | !Tube !!conc of miniprep ng/ul total=(100ul) !! 1uL miniprep added to x uL milliQ |
|- | |- | ||
| - | + | |5A || 141 || 522 | |
|- | |- | ||
| - | + | |6A ||212 || 784 | |
|- | |- | ||
| - | + | |11A ||221 || 817 | |
|- | |- | ||
| - | + | |10B || 253 ||936 | |
| + | |} | ||
| + | |||
| + | ==Site dirrected Mutagenesis Round #2 == | ||
| + | *Applied the [[Melbourne/Site directed mutagenesis|stratagene Site directed mutagenesis protocol]]to the following DNA and primer pairs, which when plated out on LB AMP plates producing the numbers of colonies shown. Several of these were picked and tubes marked with a character suffix as shown in table. | ||
| + | |||
| + | Changed PCR conditions on some primers that didn't work before to see if they would work. Specifically segment1 increased from 1 minute @ 95deg to 2minutes, segmet 2 increased from 50 sec @ 95 deg to 1 minute, and elongation time increased from 10 min @ 68 deg to 15 minutes. | ||
| + | {| border="1" | ||
| + | |+ Site dirrected Mutagenesis round #2 | ||
|- | |- | ||
| - | ! | + | !Mutation Number !!Template DNA (10ng)!! Sence Primer !! Antisence Primer !!#Colonies !! Picks named |
| - | + | ||
| - | ! | + | |
| - | + | ||
| - | ! | + | |
|- | |- | ||
| - | ! | + | !21|| 5A|| GvpL-g351a || GvpL-g351a-R || 21A,21B|| |
|- | |- | ||
| - | ! | + | !22|| 6A || GvpL-g318a || GvpL-g318a-R || 3|| 22A,22B,22C |
|- | |- | ||
| - | ! | + | !23|| 11A || GvpQ-g183a || GvpQ-g183a-R || 26|| 23A,23B |
|- | |- | ||
| - | ! | + | !24|| 10B || GvpP-g441a || GvpP-g441a-R || 60|| 24A,24B |
|- | |- | ||
| - | ! | + | !25|| 6A || GvpL-g696a || GvpL-g696a-R || 1|| 25A,25B (new PCR conditions) |
|- | |- | ||
| - | ! | + | !26|| 6A || GvpL-g213a || GvpL-g213a-R || || 26A,26B (new PCR conditions) |
|- | |- | ||
| - | ! | + | !27|| 10B || GvpQ-g150a || GvpQ-g150a-R || || 27A,27B (new PCR conditions) |
|- | |- | ||
| - | ! | + | !28|| 10B || GvpQ-g150a || GvpQ-g150a-R || None|| |
|} | |} | ||
| + | *Ran gel of 10uL sample of pcr reaction with 1uL of bluejuice. | ||
| + | ** showed lane GvpQ-g150a non functional in old conditions but functional in new conditions. | ||
* Picked colonies from Plates and [[Melb:Growing up cells|culture overnight ]] | * Picked colonies from Plates and [[Melb:Growing up cells|culture overnight ]] | ||
* [[Melbourne/Glycerol Stocks|Produced glycerol stocks]] from 900uL of each overnight culture. | * [[Melbourne/Glycerol Stocks|Produced glycerol stocks]] from 900uL of each overnight culture. | ||
Revision as of 13:34, 25 October 2007
Prepared for site dirrected mutagenesis
- Produced NZY broth 100mL autoclaved (due to contamination in last batch).
- Diluted DNA from last round
| Tube | conc of miniprep ng/ul total=(100ul) | 1uL miniprep added to x uL milliQ |
|---|---|---|
| 5A | 141 | 522 |
| 6A | 212 | 784 |
| 11A | 221 | 817 |
| 10B | 253 | 936 |
Site dirrected Mutagenesis Round #2
- Applied the stratagene Site directed mutagenesis protocolto the following DNA and primer pairs, which when plated out on LB AMP plates producing the numbers of colonies shown. Several of these were picked and tubes marked with a character suffix as shown in table.
Changed PCR conditions on some primers that didn't work before to see if they would work. Specifically segment1 increased from 1 minute @ 95deg to 2minutes, segmet 2 increased from 50 sec @ 95 deg to 1 minute, and elongation time increased from 10 min @ 68 deg to 15 minutes.
| Mutation Number | Template DNA (10ng) | Sence Primer | Antisence Primer | #Colonies | Picks named |
|---|---|---|---|---|---|
| 21 | 5A | GvpL-g351a | GvpL-g351a-R | 21A,21B | |
| 22 | 6A | GvpL-g318a | GvpL-g318a-R | 3 | 22A,22B,22C |
| 23 | 11A | GvpQ-g183a | GvpQ-g183a-R | 26 | 23A,23B |
| 24 | 10B | GvpP-g441a | GvpP-g441a-R | 60 | 24A,24B |
| 25 | 6A | GvpL-g696a | GvpL-g696a-R | 1 | 25A,25B (new PCR conditions) |
| 26 | 6A | GvpL-g213a | GvpL-g213a-R | 26A,26B (new PCR conditions) | |
| 27 | 10B | GvpQ-g150a | GvpQ-g150a-R | 27A,27B (new PCR conditions) | |
| 28 | 10B | GvpQ-g150a | GvpQ-g150a-R | None |
- Ran gel of 10uL sample of pcr reaction with 1uL of bluejuice.
- showed lane GvpQ-g150a non functional in old conditions but functional in new conditions.
- Picked colonies from Plates and culture overnight
- Produced glycerol stocks from 900uL of each overnight culture.
- Minipreped remaining culture (4mL) giving 100uL.
- Measure DNA concentrations using 2uL of miniprep(100uL) diluted in 98uL of TE.
| History | Mutation tube\colony: | A | B | C | D | E |
|---|---|---|---|---|---|---|
| pNL29T1-(GvpP-g441a)-> | 3 | 141 | 112 | |||
| pNL29T1-(GvpL-g318a)-> | 4 | 108 | 218 | |||
| pNL29T1-(Comp-g318a)-> | 5 | 141 | 119 | |||
| pNL29T1-(GvpL-g351a)-> | 6 | 212 | ||||
| pNL26P3-(GvpQ-g183a)-> | 10 | 305 | 253 | 258 | 392 | 272 |
| pNL26P3-(GvpP-g441a)-> | 11 | 221 | 190 | 259 | 204 |
- Diagnostic digests were performed using
- A) 21.5uL of each in Buffer3 with 1uL PstI (20U) at 37degC for 2h30min. (25uL reaction)
- B) 21.5uL of each p29T1 based colony in Buffer2 with 1uL EcoRI (20U) and 1uL BamHI (20U) at 37degC for 2h30min. (26uL reaction)
- C) 21.5uL of each pNL26P3 based colony and in Buffer2 with 1uL EcoRI (20U) at 37degC for 2h30min. (25uL reaction)
- Prepared two 20 lane 100mL agarose gel
- Loaded 20uL of digest
- Digest Pattern
- Expected effects
| History | Mutation tube | FRAGMENTS | |||||||
|---|---|---|---|---|---|---|---|---|---|
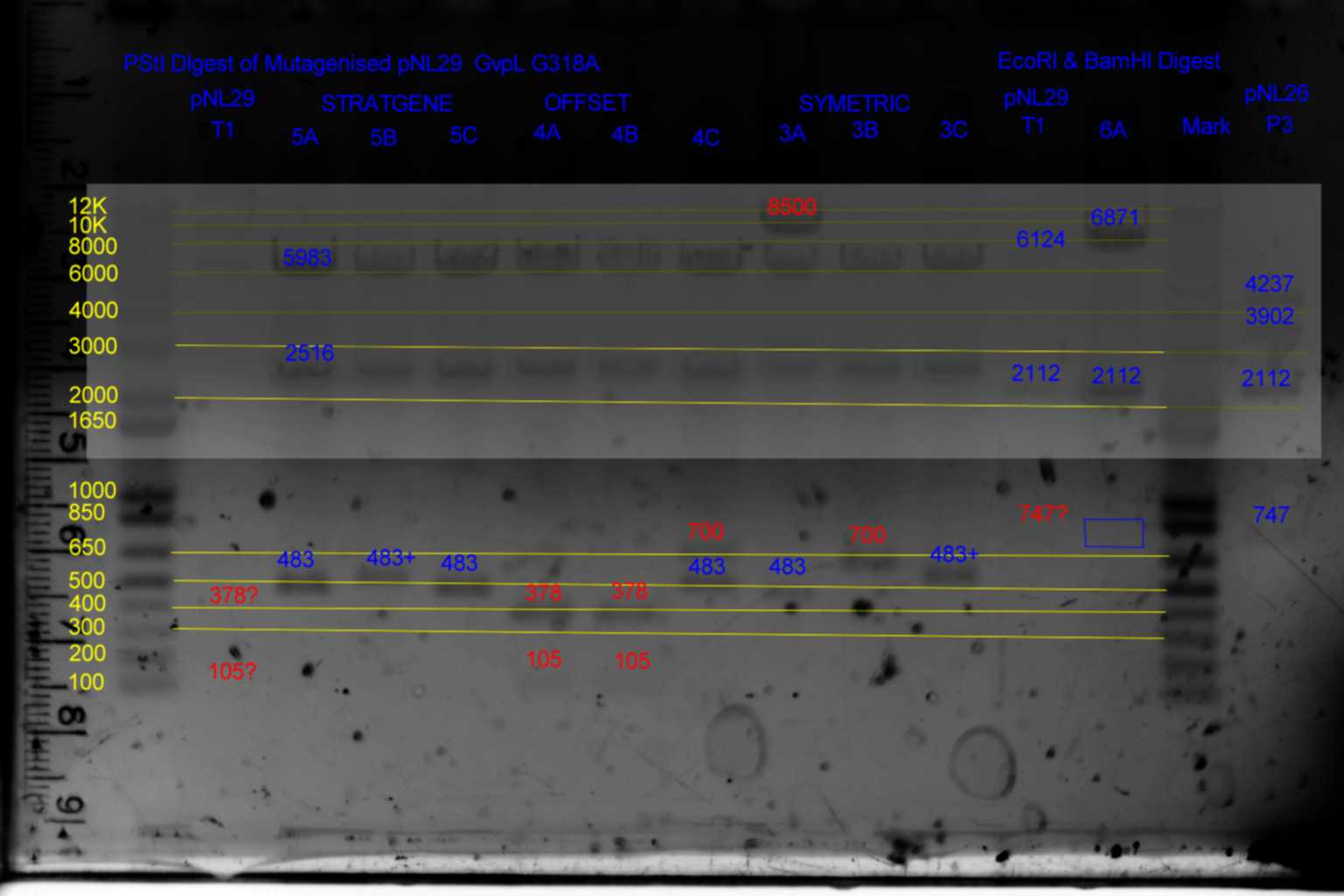
| pNL29T1 | pNL29T1 | 5983 | 2516 | 378 | 105 | ||||
| pNL29T1-(GvpP-g441a)-> | 3 | 6088 | 2516 | 378 | <- | ||||
| pNL29T1-(GvpL-g318a)-> | 4 | 5983 | 2516 | 483 | <- | ||||
| pNL29T1-(Comp-g318a)-> | 5 | 5983 | 2516 | 483 | <- | ||||
| pNL29T1-(GvpL-g351a)-> | 6 | 5983 | 2516 | 378 | 105 | ||||
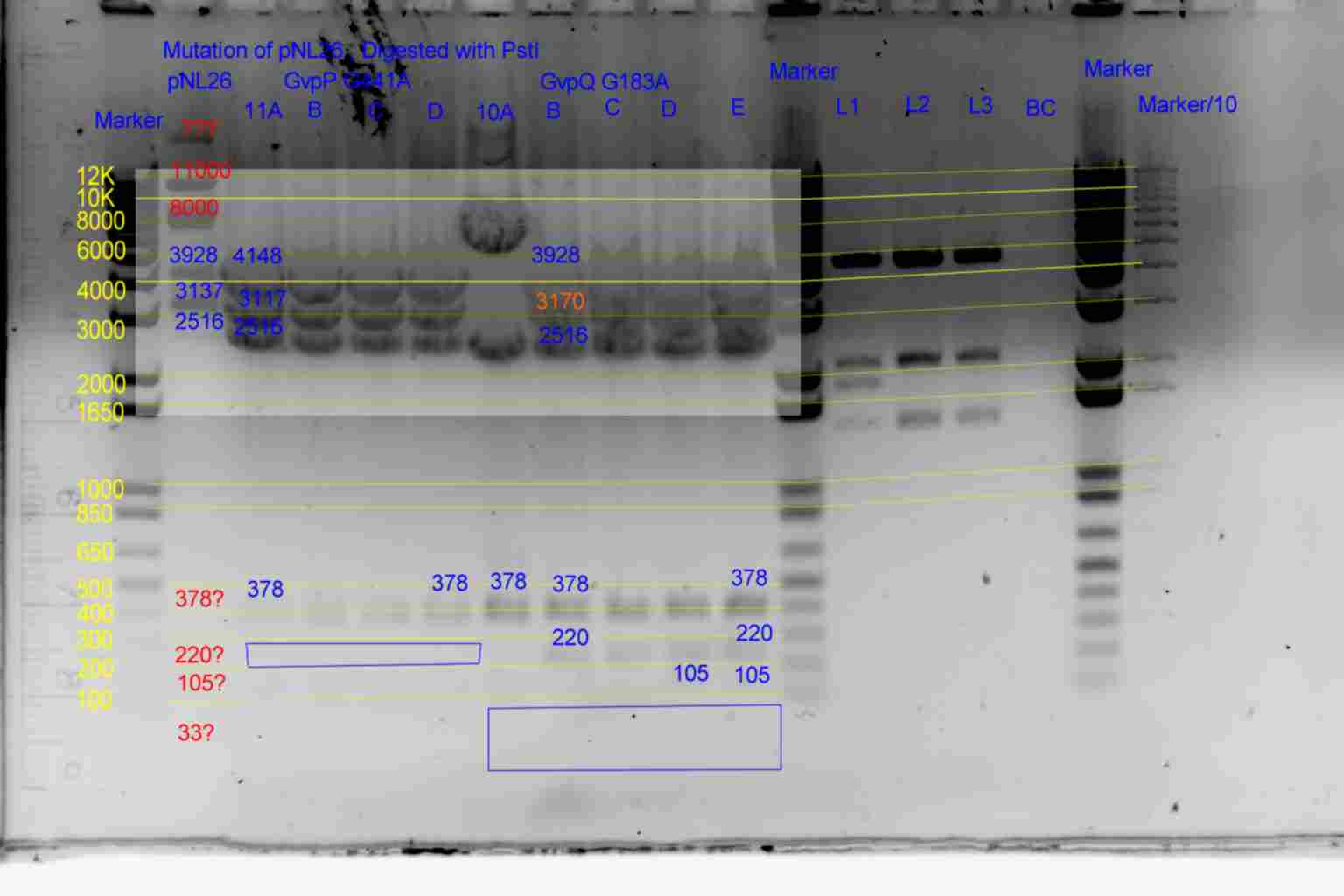
| pNL26P3 | pNL26P3 | 3928 | 3137 | 2516 | 378 | 220 | 105 | 33 | |
| p26P3-(GvpQ-g183a)-> | 10 | 3928 | 3170 | 2516 | 378 | 220 | 105 | <- | |
| p26P3-(GvpP-g441a)-> | 11 | 4148 | 3137 | 2516 | 378 | <- | 105 | 33 |
| History | Mutation tube | Fragments: | |||
|---|---|---|---|---|---|
| pNL29T1 | pNL29T1 | 6124 | 2112 | 747 | |
| pNL29T1-(GvpP-g441a)-> | 3 | 6124 | 2112 | 747 | |
| pNL29T1-(GvpL-g318a)-> | 4 | 6124 | 2112 | 747 | |
| pNL29T1-(Comp-g318a)-> | 5 | 6124 | 2112 | 747 | |
| pNL29T1-(GvpL-g351a)-> | 6 | 6871 | 2112 | <- |
- Results of PstI diagnostics:
- pNL29 control was barely visable.
- Bands less than 300bp were not visable
- Colonies 5A,5B,5C,and 3C all showed bands at 483bp and not at 378bp as required although 5B and 3C appear slightly larger than the other two.
- Colony 6A shows no band at 747bp and a band at 6871bp as required.
- Colonies 4A,and 4B contain a band at 378bp indicating failure of mutagenesis.
- The following colonies show additional bands of unknown origin: 3B and 4C (700bp) , 3A (8500bp) of unknown origin.
- See Gel
- Conclude that 5A and 6A are suitable templates for the second round of mutagenesis.
- Results of EcoRI diagnostics:
- pNL26 control barely visable.
- Colonies 11A,B,C,D and 10A,B,C,D,E all show required pattern but the 33bp band is unlikely to be visible making the 10A..E test inconclusive, and the 220bp band in 11A..D is only barely visible in 11A..E making its diagnosis also risky.
- See Gel
- Conclude that 10B and 11A may be suitable templates for the second round of mutagenesis.
- Conclude that Primers generated by the stratagene program Comp-g318a outperformed (60 colonies of which 2/3 provided correct product ) both offset primers JMY-g318a (26 colonies of which 2/3 gave template DNA)primers and hand designed primers Gvp-g318a (3 colonies of which when using the stratagene PCR protocol.
| Primers name | Design protocol | no colonies | #correctly mutated | #unknown | # template |
|---|---|---|---|---|---|
| (Comp-g318a) | Statagene | 60 | 2 | 1 | - |
| (JMY-g318a) | Offset | 26 | - | 1 | 2 |
| (GvpL-g318a) | Hand design | 3 | - | 3 | - |
- It was however also noted that the success of the PCR with the hand generated primers was negatively correlated with the melting temperature of the primers.
- It was therefore decided to increase the initial melting time from 1 to 2 minutes and increase subsequent melting times from 50 seconds to 1 minute in the next round of mutagenesis.