Melbourne/Lab Notebook gv 5
From 2007.igem.org
< Melbourne(Difference between revisions)
| Line 20: | Line 20: | ||
{| border="1" | {| border="1" | ||
| - | |+ Site | + | |+ Site directed Mutagenesis round #3 |
|- | |- | ||
!Mutation Number !!Template DNA (10ng)!! Sence Primer !! Antisence Primer !!#Colonies !! Picks named | !Mutation Number !!Template DNA (10ng)!! Sence Primer !! Antisence Primer !!#Colonies !! Picks named | ||
|- | |- | ||
| - | ! | + | !41|| 31A|| GvpL-g213a || GvpL-g213a-R || lots || 41A,41B,41C |
|- | |- | ||
| - | ! | + | !42|| 42A|| GvpL-g213a || GvpL-g213a-R || lots || 42A,42B |
|- | |- | ||
| - | ! | + | !43|| 32B|| GvpL-g696a || GvpL-g696a-R || lots || 43A,43B |
|- | |- | ||
| - | ! | + | !44|| 33A || GvpA-t57c || GvpA-t57c-R || lots || 44A,44B |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| - | + | ||
| - | + | ||
* Picked colonies from Plates and [[Melb:Growing up cells|culture overnight ]] | * Picked colonies from Plates and [[Melb:Growing up cells|culture overnight ]] | ||
* [[Melbourne/Glycerol Stocks|Produced glycerol stocks]] from 900uL of each overnight culture. | * [[Melbourne/Glycerol Stocks|Produced glycerol stocks]] from 900uL of each overnight culture. | ||
| Line 50: | Line 41: | ||
|+ DNA concentrations in ng/uL | |+ DNA concentrations in ng/uL | ||
|- | |- | ||
| - | !History!!Mutation tube\colony: !! A !! B !!C | + | !History!!Mutation tube\colony: !! A !! B !! C |
|- | |- | ||
| - | !pNL29T1-(Comp-g318a)-(GvpL-g351a)->!! | + | !pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)->!!41|| 111 || 115 || 129 |
|- | |- | ||
| - | !pNL29T1-(GvpL-g351a)-(GvpL- | + | !pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)->!!42|| 133 || 129 |
|- | |- | ||
| - | ! | + | !pNL29T1-(GvpL-g351a)-(GvpL-g318a))-(GvpL-g213a)-(GvpL-g696a)->!!43|| 107 || 75 |
|- | |- | ||
| - | !pNL26P3 | + | !pNL26P3-(GvpP-g441a)-(GvpQ-g183a))-(GvpQ-g150a)-(GvpA-t57c)->!!44|| 221 || 204 |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
* Diagnostic digests were performed using | * Diagnostic digests were performed using | ||
| - | **A) 21.5uL of each in Buffer3 with 1uL PstI (20U) at 37degC for | + | **A) 21.5uL of each in Buffer3 with 1uL PstI (20U) at 37degC for 2hrs. (25uL reaction) |
| - | **B) | + | **B) 5uL of each in Buffer2 with 1uL EcoRI (20U) and 1uL BamHI (20U) with 16.5uL of milliQ at 37degC for 2h. (26uL reaction) except pNL26 based 44A,B which were digested with EcoRI only. |
| - | **C) | + | **C) 10 uL of each in Buffer2 with 2uL HindIII (20U) with 11.5 uL milliQ at 37degC for 1hour. (25uL reaction) |
| - | * Prepared | + | * Prepared one 2row x 20 lane gel 100mL [[Melbourne/Preparing an agarose gel|agarose gel]] |
*[[Melbourne/Loading a DNA gel|Loaded]] 20uL of digest | *[[Melbourne/Loading a DNA gel|Loaded]] 20uL of digest | ||
*Digest Pattern | *Digest Pattern | ||
| Line 79: | Line 64: | ||
!History!!Mutation tube !!FRAGMENTS | !History!!Mutation tube !!FRAGMENTS | ||
|- | |- | ||
| - | !pNL29T1-(Comp-g318a)-(GvpL-g351a)->!! | + | !pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)->!!41|| !! 8982!! <- |
|- | |- | ||
| - | !pNL29T1-(GvpL-g351a)-(GvpL- | + | !pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)->!!42|| !! 8982!! <- |
|- | |- | ||
| - | ! | + | !pNL29T1-(GvpL-g351a)-(GvpL-g318a))-(GvpL-g213a)-(GvpL-g696a)->!!43|| !! 8982!! <- |
|- | |- | ||
| - | !pNL26P3-(GvpQ-g183a)-( | + | !pNL26P3-(GvpP-g441a)-(GvpQ-g183a))-(GvpQ-g150a)-(GvpA-t57c)->!!44|| !! 7318 || 2516 || 378 || 105 |
| + | |} | ||
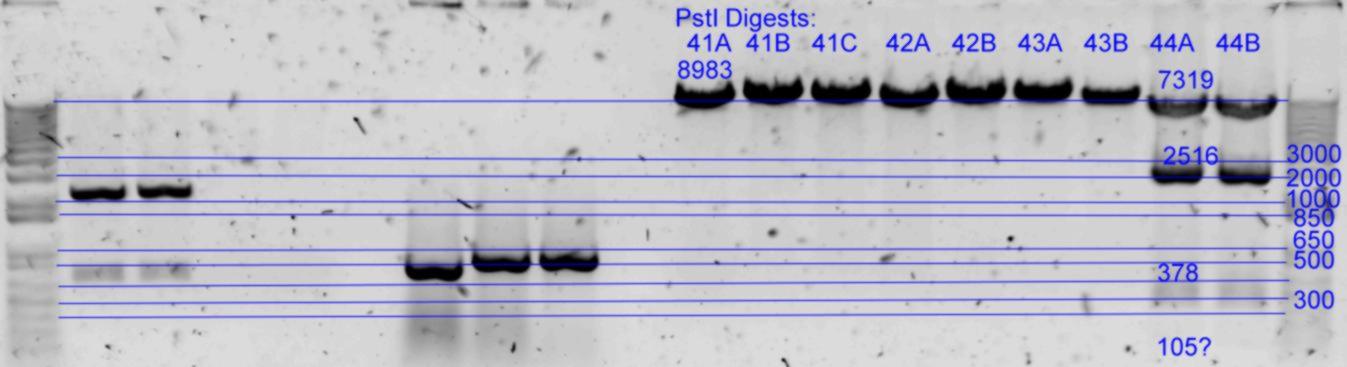
| + | *Results of PstI diagnostics:[[Image:Melbourne-gv mut 4p.jpg|left|thumb|850px|PstI digest]] | ||
| + | |||
| + | {| border="1" | ||
| + | |+ EcoRI and BamHI digest (44 with EcoRI only) | ||
|- | |- | ||
| - | ! | + | !History!!Mutation tube !!FRAGMENTS |
|- | |- | ||
| - | !pNL29T1-(GvpL-g351a)-(GvpL-g213a)->!! | + | !pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)->!!41|| || 6871|| 2112 |
|- | |- | ||
| - | ! | + | !pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)->!!42|| || 6871|| 2112 |
| + | |- | ||
| + | !pNL29T1-(GvpL-g351a)-(GvpL-g318a))-(GvpL-g213a)-(GvpL-g696a)->!!43|| || 6871|| 2112 | ||
| + | |- | ||
| + | !pNL26P3-(GvpP-g441a)-(GvpQ-g183a))-(GvpQ-g150a)-(GvpA-t57c)->!!44 || !! 10318 || <- | ||
|} | |} | ||
| - | + | ||
| - | + | ||
{| border="1" | {| border="1" | ||
| - | |+ | + | |+ HindIII digest |
|- | |- | ||
| - | !History!!Mutation tube!! | + | !History!!Mutation tube !!FRAGMENTS |
|- | |- | ||
| - | !pNL29T1-(Comp-g318a)-(GvpL-g351a)->!! | + | !pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)->!!41|| || 4852|| 3479 || 652 |
|- | |- | ||
| - | !pNL29T1-(GvpL-g351a)-(GvpL- | + | !pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)->!!42|| || 4852|| 3479 || 652 |
|- | |- | ||
| - | !pNL29T1-(GvpL-g351a)-(GvpL-g696a)->!! | + | !pNL29T1-(GvpL-g351a)-(GvpL-g318a))-(GvpL-g213a)-(GvpL-g696a)->!!43|| || 4852|| 3479 || 652 |
|- | |- | ||
| - | ! | + | !pNL26P3-(GvpP-g441a)-(GvpQ-g183a))-(GvpQ-g150a)-(GvpA-t57c)->!!44 || || 4852|| 3479 || 1335 || 652 |
|} | |} | ||
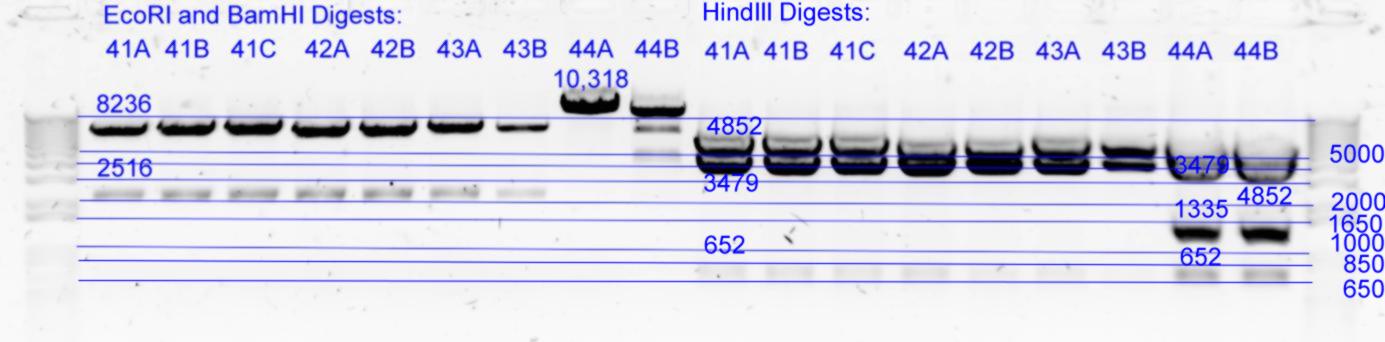
| - | + | [[Image:Melbourne-gv mut 4e.jpg|left|thumb|850px|EcoRI / BamHI digests and HindIII digest]] | |
| - | *Results of | + | *Results of digests: All look good except 44B which is still being cut ecoRI at t57c (background incomplete digestion of template using dpnI). There does however appear to be a small variation in size but it is hard to tell if this is gel or concentration inconsistancy. 41C HindII 3479 band appears slightly larger than 41A or B. |
| - | + | ||
Latest revision as of 02:40, 26 October 2007
Prepared for site dirrected mutagenesis
- Diluted DNA from last round, creating 10ng/ul sample and then adding 1uL of this to 36uL milliQ water to produce required 10ng/37uL template for PCR.
| Tube | conc of miniprep ng/ul total=(100ul) | 1uL miniprep added to x uL milliQ -> 10ng/uL |
|---|---|---|
| 31A | 117 | 10.7 |
| 31B | 133 | 12.3 |
| 32B | 96 | 8.6 |
| 33A | 114 | 10.4 |
Site dirrected Mutagenesis Round #4
- Applied the stratagene Site directed mutagenesis protocolto the following DNA and primer pairs, which when plated out on LB AMP plates producing the numbers of colonies shown. Several of these were picked and tubes marked with a character suffix as shown in table.
| Mutation Number | Template DNA (10ng) | Sence Primer | Antisence Primer | #Colonies | Picks named |
|---|---|---|---|---|---|
| 41 | 31A | GvpL-g213a | GvpL-g213a-R | lots | 41A,41B,41C |
| 42 | 42A | GvpL-g213a | GvpL-g213a-R | lots | 42A,42B |
| 43 | 32B | GvpL-g696a | GvpL-g696a-R | lots | 43A,43B |
| 44 | 33A | GvpA-t57c | GvpA-t57c-R | lots | 44A,44B |
- Picked colonies from Plates and culture overnight
- Produced glycerol stocks from 900uL of each overnight culture.
- Minipreped remaining culture (4mL) giving 100uL.
- Measure DNA concentrations using 5uL of miniprep(100uL) diluted in 45uL of TE.
| History | Mutation tube\colony: | A | B | C |
|---|---|---|---|---|
| pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)-> | 41 | 111 | 115 | 129 |
| pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)-> | 42 | 133 | 129 | |
| pNL29T1-(GvpL-g351a)-(GvpL-g318a))-(GvpL-g213a)-(GvpL-g696a)-> | 43 | 107 | 75 | |
| pNL26P3-(GvpP-g441a)-(GvpQ-g183a))-(GvpQ-g150a)-(GvpA-t57c)-> | 44 | 221 | 204 |
- Diagnostic digests were performed using
- A) 21.5uL of each in Buffer3 with 1uL PstI (20U) at 37degC for 2hrs. (25uL reaction)
- B) 5uL of each in Buffer2 with 1uL EcoRI (20U) and 1uL BamHI (20U) with 16.5uL of milliQ at 37degC for 2h. (26uL reaction) except pNL26 based 44A,B which were digested with EcoRI only.
- C) 10 uL of each in Buffer2 with 2uL HindIII (20U) with 11.5 uL milliQ at 37degC for 1hour. (25uL reaction)
- Prepared one 2row x 20 lane gel 100mL agarose gel
- Loaded 20uL of digest
- Digest Pattern
- Expected effects
| History | Mutation tube | FRAGMENTS | ||||
|---|---|---|---|---|---|---|
| pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)-> | 41 | 8982 | <- | |||
| pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)-> | 42 | 8982 | <- | |||
| pNL29T1-(GvpL-g351a)-(GvpL-g318a))-(GvpL-g213a)-(GvpL-g696a)-> | 43 | 8982 | <- | |||
| pNL26P3-(GvpP-g441a)-(GvpQ-g183a))-(GvpQ-g150a)-(GvpA-t57c)-> | 44 | 7318 | 2516 | 378 | 105 |
- Results of PstI diagnostics:
| History | Mutation tube | FRAGMENTS | ||
|---|---|---|---|---|
| pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)-> | 41 | 6871 | 2112 | |
| pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)-> | 42 | 6871 | 2112 | |
| pNL29T1-(GvpL-g351a)-(GvpL-g318a))-(GvpL-g213a)-(GvpL-g696a)-> | 43 | 6871 | 2112 | |
| pNL26P3-(GvpP-g441a)-(GvpQ-g183a))-(GvpQ-g150a)-(GvpA-t57c)-> | 44 | 10318 | <- |
| History | Mutation tube | FRAGMENTS | ||||
|---|---|---|---|---|---|---|
| pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)-> | 41 | 4852 | 3479 | 652 | ||
| pNL29T1-(Comp-g318a)-(GvpL-g351a))-(GvpL-g696a)-(GvpL-g213a)-> | 42 | 4852 | 3479 | 652 | ||
| pNL29T1-(GvpL-g351a)-(GvpL-g318a))-(GvpL-g213a)-(GvpL-g696a)-> | 43 | 4852 | 3479 | 652 | ||
| pNL26P3-(GvpP-g441a)-(GvpQ-g183a))-(GvpQ-g150a)-(GvpA-t57c)-> | 44 | 4852 | 3479 | 1335 | 652 |
- Results of digests: All look good except 44B which is still being cut ecoRI at t57c (background incomplete digestion of template using dpnI). There does however appear to be a small variation in size but it is hard to tell if this is gel or concentration inconsistancy. 41C HindII 3479 band appears slightly larger than 41A or B.