Imperial/Cell by Date/Testing
From 2007.igem.org
m |
m (→Validation and Conclusion) |
||
| Line 60: | Line 60: | ||
|} | |} | ||
<br clear="all"> | <br clear="all"> | ||
| + | |||
| + | |||
| + | Previous work has been carried out to model the spoilage of ground beef by living organisms. | ||
| + | Above in figure 1 is is a model developed by Koutsoumanis in 2006 which colesly fit the behaviour of the spoilge organism we are interested in, Pseudomonas, under dynamic temperature conditions. One of the Key conclusions drawn from this model is the almost instantaneous repsone time of the Pseudomonas' growth parameter. The result of this is that our system needs to have a quick respone time to correclty report the temperature history of the beef. | ||
| + | |||
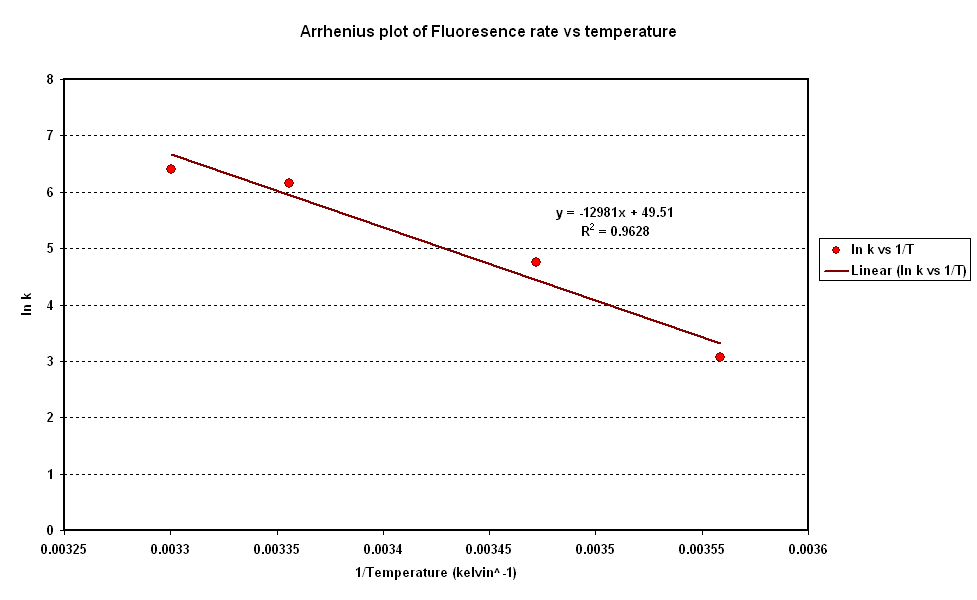
| + | Koutsoumanis' and also one Giannuzzi developed in 1998 are both based on the Gompertz model. This model allows some insight into the mechanisms of ground beef spoilage. In particular through manipulation of the Gompertz Parameters and assuming a Arrhenius type relationship between Pseudomonas' growth parameter and temperature we can infer the Activation energy of the spoilage reaction. This is shown in figure 2 in which a stongly linear behaviour allows us to continue with our Arrhenius assumption and extract that Activatoin energy of the spoilge reaction. As given in our specifications the Activation energy for ground meat seem to be around 30kJ/mol. The result of this as per Taoukis' work is that our system needs to have a similar activation energy. I hope to determine our system activation energy in the same way Giannuzzi determined Pseudomonas'. | ||
Revision as of 13:34, 22 October 2007

Testing and Discussion
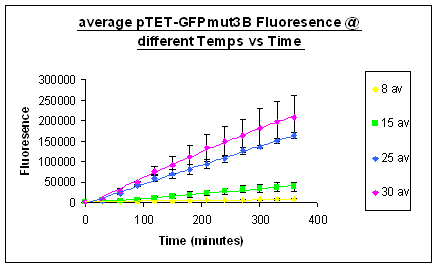
Investigation properties of system under isothermal scenarios
Previous work has been carried out to model the spoilage of ground beef by living organisms. Above in figure 1 is is a model developed by Koutsoumanis in 2006 which colesly fit the behaviour of the spoilge organism we are interested in, Pseudomonas, under dynamic temperature conditions. One of the Key conclusions drawn from this model is the almost instantaneous repsone time of the Pseudomonas' growth parameter. The result of this is that our system needs to have a quick respone time to correclty report the temperature history of the beef.
Koutsoumanis' and also one Giannuzzi developed in 1998 are both based on the Gompertz model. This model allows some insight into the mechanisms of ground beef spoilage. In particular through manipulation of the Gompertz Parameters and assuming a Arrhenius type relationship between Pseudomonas' growth parameter and temperature we can infer the Activation energy of the spoilage reaction. This is shown in figure 2 in which a stongly linear behaviour allows us to continue with our Arrhenius assumption and extract that Activatoin energy of the spoilge reaction. As given in our specifications the Activation energy for ground meat seem to be around 30kJ/mol. The result of this as per Taoukis' work is that our system needs to have a similar activation energy. I hope to determine our system activation energy in the same way Giannuzzi determined Pseudomonas'.
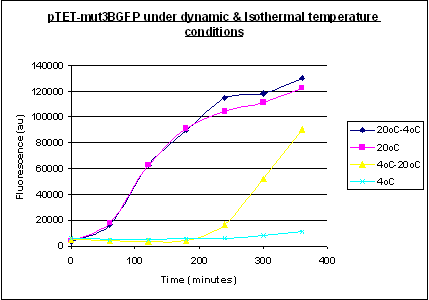
Investigation System properties under Dynamic Temperature Conditions
With some understanding of how Pseuodmonas spoils aerobically stored ground beef it is really important for us to gain some understanding of how our sytem behaves in similar scenarios to the models developed by Koutsoumanis and Giannuzi in order to extract the same parameters.
For Isothermal conditions we have been able to devlope some simple models of the gene expression of our sytem with a strong dependance on energy depletion as will feel this is the major limiting factor of our system. As seen above in figures 3 and 4 with energy depletion as our major limiting factor we expect the concentration of our reporter gene to increase to a peak and then as energy runs out expressoin is curbed and decay of our reporter takes over giving an exponential like decay.
Validation and Conclusion
| Inputs | ||
| Outputs | ||
| Activation Energy | ||
| Health Regulations | ||
| Response Time | ||
| Lifespan |
Previous work has been carried out to model the spoilage of ground beef by living organisms.
Above in figure 1 is is a model developed by Koutsoumanis in 2006 which colesly fit the behaviour of the spoilge organism we are interested in, Pseudomonas, under dynamic temperature conditions. One of the Key conclusions drawn from this model is the almost instantaneous repsone time of the Pseudomonas' growth parameter. The result of this is that our system needs to have a quick respone time to correclty report the temperature history of the beef.
Koutsoumanis' and also one Giannuzzi developed in 1998 are both based on the Gompertz model. This model allows some insight into the mechanisms of ground beef spoilage. In particular through manipulation of the Gompertz Parameters and assuming a Arrhenius type relationship between Pseudomonas' growth parameter and temperature we can infer the Activation energy of the spoilage reaction. This is shown in figure 2 in which a stongly linear behaviour allows us to continue with our Arrhenius assumption and extract that Activatoin energy of the spoilge reaction. As given in our specifications the Activation energy for ground meat seem to be around 30kJ/mol. The result of this as per Taoukis' work is that our system needs to have a similar activation energy. I hope to determine our system activation energy in the same way Giannuzzi determined Pseudomonas'.