Chiba/Project Design
From 2007.igem.org
(→How Our System Works) |
(→How Our System Works) |
||
| (60 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
[[Image:chiba_logo.png|center]] | [[Image:chiba_logo.png|center]] | ||
__NOTOC__ | __NOTOC__ | ||
| - | {| style="border:0;width:100%;" cellpadding="20px" cellspacing="0" | + | {| style="border:0;width:100%;font-family:'Trebuchet MS'" cellpadding="20px" cellspacing="0" |
| align="center" | | | align="center" | | ||
| - | [[Chiba|Introduction]] | [[Chiba/Project_Design|Project Design]] | + | [[Chiba|Home]]<br> |
| + | <span style="font-size:120%;font-weight:bold;">[[Chiba/Introduction|Introduction]] | [[Chiba/Project_Design|Project Design]] ( [[Chiba/Engeneering_Flagella|1.Affinity Tag]] | [[Chiba/Communication|2.Communication Module]] | [[Chiba/Quorum_Sensing|3.Size Control]] ) | [[Chiba/Making Marimo|Making Marimos]] | [[Chiba/Goal|Our Goal]]</span><br> | ||
| + | [[Chiba/Acknowledgements|Acknowledgements]] | [[Chiba/Team_Members|Team Members]] | [http://chem.tf.chiba-u.jp/igem/ iGEM Chiba Website] | [[Chiba/Members_Only|メンバ連絡簿]] | ||
|} | |} | ||
| Line 10: | Line 12: | ||
==Project Design== | ==Project Design== | ||
===Concept=== | ===Concept=== | ||
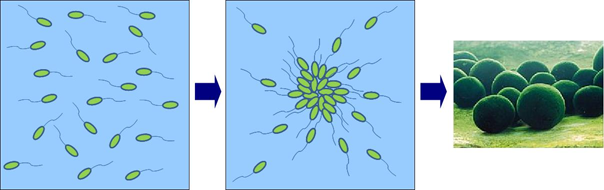
| - | + | [[Image:Story.PNG|frame|'''Fig. 4''' Concept|center]]<br> | |
| - | [[Image:Story.PNG| | + | We aimed to make a spherical gathering of bacteria such like marimo by ordering bacteria go get together and stick to each other. |
| - | + | ||
| - | === | + | ===What our system requires=== |
| - | + | {| style="border:0px;" cellpadding="5px" | |
| - | + | | width="30%" valign="top" | | |
| + | ====[[Chiba/Engeneering_Flagella|1.Affinity Tag]]==== | ||
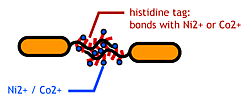
| + | Make a '''His-tagged Flagella'''. We aimed to stick bacteria by displaying histidines (which bonds each other through metal ions) on the flagellar filament.<br> | ||
| - | + | [[Image:Chiba_stickbacteria.png|frame|left|'''Fig. 5 ''' His-tagged flagella as a bactria linker. This image depicts only one tail as flagella (the real bacteria have about ten flagella per cell).]]<br> | |
| - | + | | width="30%" valign="top" | | |
| - | + | ||
| - | + | ||
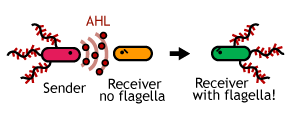
| - | + | ====[[Chiba/Communication|2.Communication Module]]==== | |
| - | + | Make a '''Bacterial Communication'''. We make 2 types of cell(sender&receiver) having different gene circuit. Senders sticking each other in advance send a signal to receivers and receivers grow affinity tags.<br> | |
| - | + | ||
| - | ====[[Chiba/ | + | |
| - | Make a ''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | ====[[Chiba/ | + | [[Image:Chiba_prj_com.png|frame|left|'''Fig. 6''' Bacterial Communication.]] |
| - | Make an '''AHL | + | | width="30%" valign="top" | |
| - | [[Image:Chiba_proj_ahlconc.png|frame|left|''' | + | |
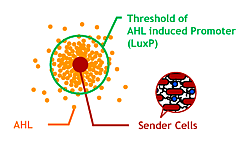
| + | ====[[Chiba/Quorum_Sensing|3.Size Control]]==== | ||
| + | Make an '''AHL localized region''' for quorum sensing. | ||
| + | [[Image:Chiba_proj_ahlconc.png|frame|left|'''Fig. 7''' Controlling AHL diffusing area and the size of Bacteria Marimo.]] | ||
|} | |} | ||
| - | === | + | ===How Our System Works=== |
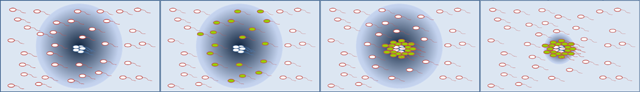
| - | [[Image: | + | [[Image:How our system works chiba.PNG|frame|center|'''Fig. 8''' How Our System Works.]]<br> |
| - | + | #Senders whose flagella display the histidine tags stick to each other through metal ions. This becomes the core of Bacteria Marimo. The senders also produce AHL to sign the receiver cells.<br> | |
| - | + | #Receivers express his/flagella and GFP in high [AHL]; only when they get close to the senders core.<br> | |
| - | [ | + | #Receivers stick to the senders core and themselves through metal ions.... one after another.<br> |
| - | + | #As seen, the cluster grows like a snowball. At the same time, the receivers degrade AHL and thus AHL diffusion space around the mixed cluster limited. By controlling the rate of AHL degradation and so on, one can define the size of such Bacteria Marimo.<br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | + | '''For other strategies to make marimo, visit the official website of [http://chem.tf.chiba-u.jp/igem/ iGEMCHIBA]''' | |
| - | [ | + | <!-- SenderやReceiverの色を対応させてみたらどう? by田代 --> |
Latest revision as of 05:36, 27 October 2007
|
Home |
Project Design
Concept
We aimed to make a spherical gathering of bacteria such like marimo by ordering bacteria go get together and stick to each other.
What our system requires
1.Affinity TagMake a His-tagged Flagella. We aimed to stick bacteria by displaying histidines (which bonds each other through metal ions) on the flagellar filament. |
2.Communication ModuleMake a Bacterial Communication. We make 2 types of cell(sender&receiver) having different gene circuit. Senders sticking each other in advance send a signal to receivers and receivers grow affinity tags. |
3.Size ControlMake an AHL localized region for quorum sensing. |
How Our System Works
- Senders whose flagella display the histidine tags stick to each other through metal ions. This becomes the core of Bacteria Marimo. The senders also produce AHL to sign the receiver cells.
- Receivers express his/flagella and GFP in high [AHL]; only when they get close to the senders core.
- Receivers stick to the senders core and themselves through metal ions.... one after another.
- As seen, the cluster grows like a snowball. At the same time, the receivers degrade AHL and thus AHL diffusion space around the mixed cluster limited. By controlling the rate of AHL degradation and so on, one can define the size of such Bacteria Marimo.
For other strategies to make marimo, visit the official website of [http://chem.tf.chiba-u.jp/igem/ iGEMCHIBA]