Imperial/Infector Detector/Testing
From 2007.igem.org
(Difference between revisions)
m (→Infector Detector: Testing) |
m (→Aims) |
||
| Line 27: | Line 27: | ||
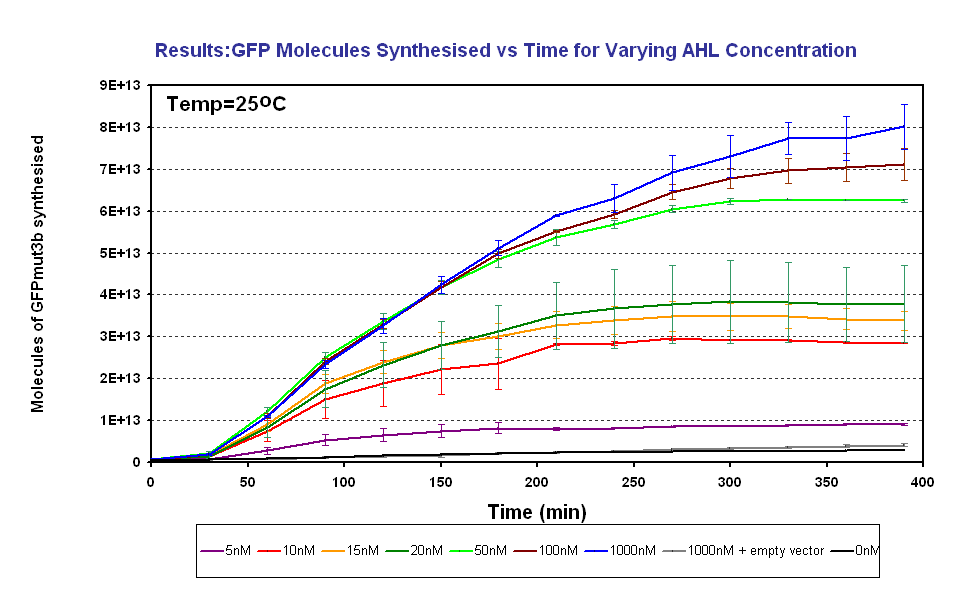
*To characterise the output of GFPmut3b for a range of AHL inputs. From this obtain the AHL sensitivity of our system. | *To characterise the output of GFPmut3b for a range of AHL inputs. From this obtain the AHL sensitivity of our system. | ||
In addition the fluorescence measurements were converted to number of GFPmut3b molecules synthesised using a calibration curve constructed using purified GFPmut3b. | In addition the fluorescence measurements were converted to number of GFPmut3b molecules synthesised using a calibration curve constructed using purified GFPmut3b. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{|align="center" | {|align="center" | ||
| - | | width="50%"|<br>[[Image:IC 2007 DNA | + | | width="50%"|<br>[[Image:IC 2007 DNA Concentration.PNG|thumb|400px|Fig.1.1:Molecules of GFPmut3b synthesised over time, for each DNA Concentration ''in vitro'' - The fluorescence was measured over time for each experiment and converted into molecules of GFPmut3b ''in vitro'' |
| - | | width="50%"|<br>[[ | + | [[Imperial/Wet Lab/Results/Res1.3/Converting_Units| using our calibration curve]].]] |
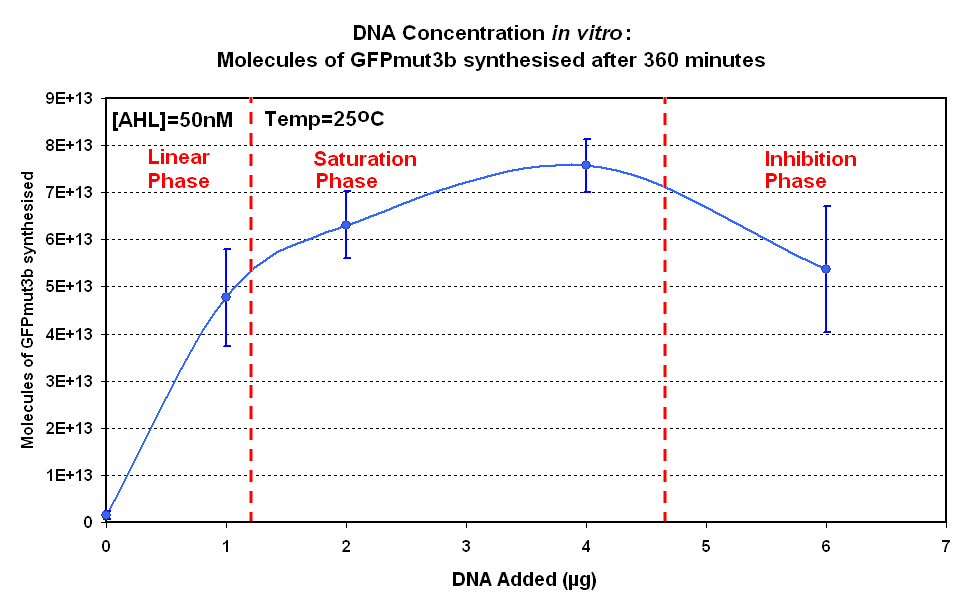
| + | | width="50%"|<br>[[image:IC 2007 DNA Concentration 360mins.PNG|thumb|450px|Fig.1.2:Molecules of GFPmut3b synthesised for each DNA Concentration ''in vitro'', after 360 minutes.]] | ||
|} | |} | ||
Revision as of 02:51, 26 October 2007

Infector Detector: Testing
Aims
The aims of the testing were as follows:
- To test and obtain the optimal DNA concentration for construct 1 in vitro
- To characterise the output of GFPmut3b for a range of AHL inputs. From this obtain the AHL sensitivity of our system.
In addition the fluorescence measurements were converted to number of GFPmut3b molecules synthesised using a calibration curve constructed using purified GFPmut3b.
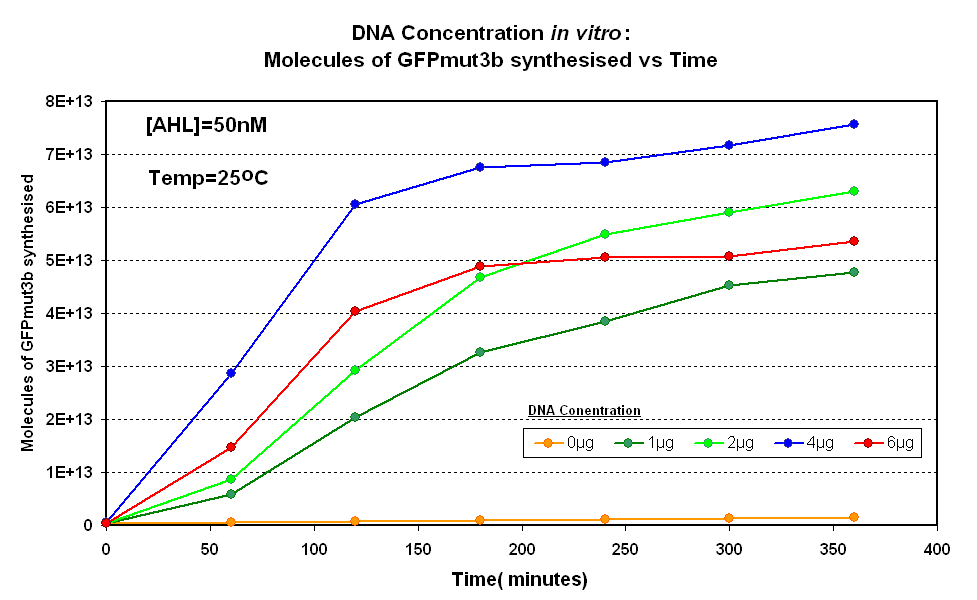 Fig.1.1:Molecules of GFPmut3b synthesised over time, for each DNA Concentration in vitro - The fluorescence was measured over time for each experiment and converted into molecules of GFPmut3b in vitro using our calibration curve. |
Results
The results show us the following:
|