Imperial/Infector Detector/F2620 Comparison
From 2007.igem.org
(→Comparison to F2620) |
m (→Comparison to F2620) |
||
| Line 45: | Line 45: | ||
<br> | <br> | ||
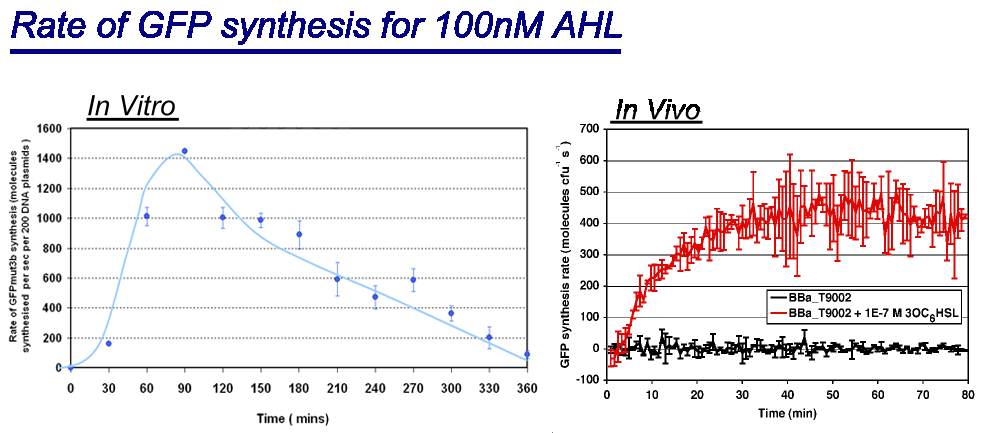
*''in vivo'' has a maximal rate of 400-500 molecules of GFP synthesised per second per cell. In addition the ''in vivo'' chassis reaches a steady state around 30minutes.<br> | *''in vivo'' has a maximal rate of 400-500 molecules of GFP synthesised per second per cell. In addition the ''in vivo'' chassis reaches a steady state around 30minutes.<br> | ||
| - | *''in vitro'' has the equivalent of 220 molecules of GFP synthesised per second per cell equivalent | + | *''in vitro'' has the equivalent of 220 molecules of GFP synthesised per second per cell equivalent, the cell equilavent being based upon the normalization of DNA plasmids. The ''in vitro'' chassis does not reach a steady state, in fact it decreases in rate of synthesis after 90 minutes and keeps decreasing until rate is zero at around 360 minutes. |
<br> | <br> | ||
| - | The reason why the ''in vitro chassis never reaches a steady state is because the ''in vitro'' chassis has limited energy and metabolites available | + | The reason why the ''in vitro chassis never reaches a steady state is because the ''in vitro'' chassis has limited energy and metabolites available this is unlike the ''in vivo'' chassis that has media available. |
<br> | <br> | ||
| - | Interestingly the values of rate of synthesis are in the same order magnitude of hundreds, this suggesting that the normalisation | + | Interestingly the values of rate of synthesis are in the same order magnitude of hundreds, this suggesting that the normalisation we are using to compare these chassis is valid. |
]] | ]] | ||
|} | |} | ||
Revision as of 23:55, 25 October 2007

Comparison to F2620
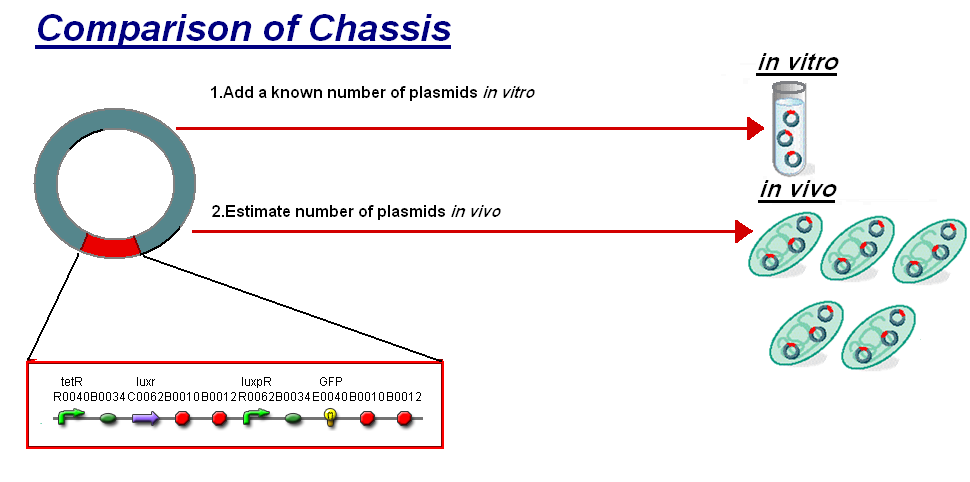
For further analysis the results of our in vitro testing have been compared to the work in vivo on [http://partsregistry.org/Part:BBa_F2620 BBa_F2620](pTet-LuxR-pLux-GFPmut3b), the construct being the same as our construct 1 for infecter detector. The motivation of the comparison is to see how this construct will respond in different chassis. To do this we investigated a standard unit s to allow the comparison between in vitro and in vivo.
The basis for comparison is to normalise the in vitro chassis on the number of plasmids to give a platform for comparison:
- In Vitro - 4µg of DNA was added which for [http://partsregistry.org/Part:BBa_T9002 pTet-LuxR-pLux-GFPmut3b] is 904823007 plasmids
- In Vivo - Each cell the plasmids number was estimated at 30 per cell
To compare we normalised the data of in vitro GFPmut3b molecules synthesised per 30 plasmids to allow some comparison to the in vivo data.
Of particular interest was to compare the:
- Rate of GFP synthesis of 100nM
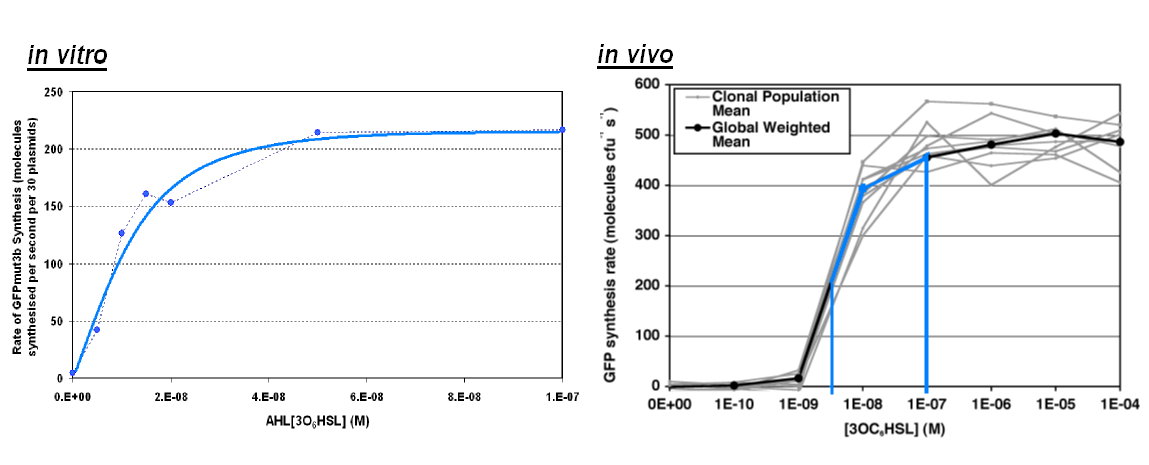
- Transfer Function.
Transfer Function
Summary
Below is list of which of the orginial Specifications that our infecter detector achieved:
| Achievements | ||
| Inputs | Sensitive to 5-1000nM | |
| Outputs | Future work - Using Stronger fluorescent protein such as DsRed express | |
| Response Time | Systems responds <30minutes | |
| Operating Conditions | System works at 25°C | |
| Health & Safety | Cell Free in vitro chassis | |
| Lifespan | Can be stored in freezer for prolonged periods | |
| Packaging | Future Work - Using our chassis in a spray |