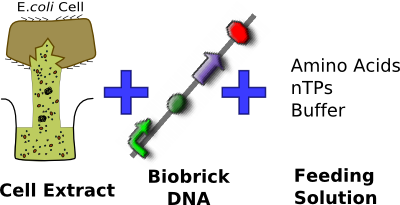Imperial/Cell-Free/Applications
From 2007.igem.org
| (17 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:IC07navmenu}} | {{Template:IC07navmenu}} | ||
__NOTOC__ | __NOTOC__ | ||
| + | <html> | ||
| + | <link rel="stylesheet" href="/igem07/index.php?title=User:Dirkvs/Stylesheets/IC07persist.css&action=raw&ctype=text/css" type="text/css" /> | ||
| - | == | + | <div id="tabs"> |
| - | + | <ul> | |
| + | <li><a href="https://2007.igem.org/Imperial/Cell-Free/Whatis" title=""><span>What is Cell-Free?</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell-Free/Comparison" title=""><span>Advantages of CFS</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell-Free/Contribution" title=""><span>Our Contributions</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell-Free/Characterisation" title=""><span>Characterisation</span></a></li> | ||
| + | <li><a class="current" href="https://2007.igem.org/Imperial/Cell-Free/Applications" title=""><span>CFS Applications</span></a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <hr /> | ||
| + | <br clear="all"> | ||
| + | </html> | ||
| - | |||
| - | |||
| + | = Cell-Free: Applications = | ||
| + | ==Our Design == | ||
| + | To get around the problem of having bacteria in contact with food or medical devices in the applications of [[Imperial/Infector_Detector/Introduction |Infector Detector]] and [[Imperial/Cell_by_Date/Introduction |Cell by Date]] respectively, we aimed to express reporter DNA constructs within a CFS consisting of various ''E. coli'' cell extracts (homemade S30, commercial S30 and commercial S30 pT7) in bulk solution. In parallel, we investigated and attempted to optimize conditions for '''phospholipid vesicle formation'''. Future work on our project would then involve combining the two, so that protein expression is achieved inside vesicles that have prolonged chassis lifespan. | ||
| + | === ''In vitro'' Expression in Bulk Solution === | ||
| + | Since we are using ''E. coli'' plasmid DNA constructs, we chose to use the compatible ''E. coli'' cell extract for in-vitro expression. We tried out four options so as to find out the system most suitable for our two applications. | ||
| - | == Cell- | + | {| border="1" |
| - | + | |- | |
| + | |style="background:#aaccff" colspan="2"| | ||
| + | <center>'''Homemade'''</center> | ||
| + | |style="background:#ffcc66" colspan="2"| | ||
| + | <center>'''Commercial'''</center> | ||
| + | |- | ||
| + | |style="background:#cceeff"|<center>'''''E. coli'' S30 extract'''</center> | ||
| + | |style="background:#cceeff"|<center>'''''E. coli'' S12 extract'''</center> | ||
| + | |style="background:#ffeebb"|<center>'''''E. coli'' S30 extract'''</center> | ||
| + | |style="background:#ffeebb"|<center>'''''E. coli'' T7 S30 extract'''</center> | ||
| + | |- | ||
| + | |colspan="2" width=50%| | ||
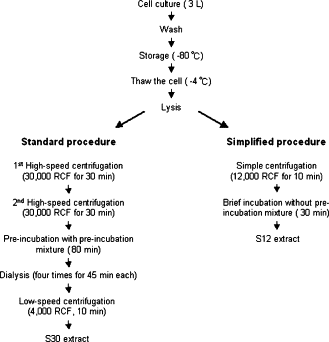
| + | *Prepared using the protocols described by Kim DM et al. (2006) <sup>[[#References |1]]</sup> | ||
| + | *Used ''E. coli'' strain BL21 (DE3) with loss of function mutations in OmpT endoproteinase and lon protease genes | ||
| + | *BL21 can express T7 RNA polymerase under the control of LacI, inducible by IPTG | ||
| + | |||
| + | [[Image:IC07_CFS-homemade.gif|center|350px]] | ||
| + | |colspan="2" width=50% valign="top"| | ||
| + | *Purchased from [http://www.promega.com/search/Default.aspx?query=s30+extract Promega] | ||
| + | *Prepared by modifications of the method described by Zubay et al. (1980) <sup>[[#References |2]]</sup> | ||
| + | *Used ''E. coli'' strain B deficient in OmpT endoproteinase and lon protease activity<bR><br><br><br><br><br><br> | ||
| + | [[Image:IC07_CFS_components.png|center|350px]] | ||
| + | |} | ||
| + | <br clear="all"> | ||
| + | |||
| + | [[Imperial/Wet_Lab/Protocols/CE1.1 | Preparation of Homemade S30 Cell Extract Protocol ]]<br> | ||
| + | [[Imperial/Wet_Lab/Protocols/CE1.2 | Preparation of Homemade S12 Cell Extract Protocol ]]<br> | ||
| + | [[Imperial/Wet_Lab/Protocols/CE1.4 | Preparation of Homemade S30 Reaction Mixture ]]<br> | ||
| + | [[Imperial/Wet_Lab/Protocols/CE1.3 | Preparation of Commercially Available S30 Reaction Mixture ]]<br> | ||
| + | |||
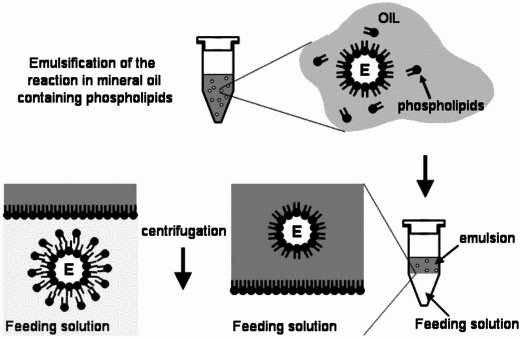
| + | === Vesicle Formation === | ||
| + | For the formation of phospholipid bilayer vesicles, we chose to use the '''mineral oil method'''. The protocols are based on Engineering Asymmetric Vesicles by Sophie Pautot, BJ Frisken, and DA Weitz. (2003) <sup>[[#References |3]]</sup> and Vincent Noireaux et al. (2005). <sup>[[#References |4]]</sup> <br><br> | ||
| + | |||
| + | [[Image:IC07_CFSvesicle.jpg|thumb|right|310px|Toward an artificial cell based on gene expression in vesicles by Vincent Noireaux et al. (2005)]] | ||
| + | |||
| + | |||
| + | The mineral oil method consists of three stages, regardless of composition or equipment: | ||
| + | #Preparation of lipid-oil suspension | ||
| + | #Emulsification of vesicle contents | ||
| + | #Bilayer formation through sedimentation | ||
| + | |||
| + | [[Imperial/Wet_Lab/Protocols/VF1.1| Vesicle Formation Protocol]] | ||
| + | |||
| + | <br clear="all"> | ||
| + | |||
| + | |||
| + | == Cell-Free in Infector Detector == | ||
| + | For more details on how [[Imperial/Infector_Detector/Introduction |Infector Detector]] integrates CFS into its system, go to its [[Imperial/Infector_Detector/Design |Design]] page. | ||
| + | |||
| + | |||
| + | == Cell-Free in Cell by Date == | ||
| + | For more details on how [[Imperial/Cell_by_Date/Introduction |Cell by Date]] utlizes CFS, go to its [[Imperial/Cell_by_Date/Design |Design]] page. For more details on the determination of CFS capabilities and characterizing it, go to its [[Imperial/Cell_by_Date/Testing |Testing/Validation]] page. | ||
| + | |||
| + | |||
| + | |||
| + | <center> [https://2007.igem.org/Imperial/Cell-Free/Characterisation<< Characterisation] | Applications | [https://2007.igem.org/Imperial Home >>] | ||
| + | </center> | ||
| + | |||
| + | == References == | ||
| + | #Kim TW, Keum JW, Oh IS, Choi CY, Park CG, and Kim DM. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J Biotechnol 2006 Dec 1; 126(4) 554-61. doi:10.1016/j.jbiotec.2006.05.014 pmid:16797767. | ||
| + | #[http://www.promega.com/guides/ive_guide/default.htm In vitro protein expression (Promega)] | ||
| + | # Sophie Pautot, B. J. Frisken and D.A. Weitz: Engineering asymmetric vesicles. Proceedings of the National Academy of Sciences 100, p. 10718-10721 (2003). | ||
| + | # Noireaux V, Bar-Ziv R, Godefroy J, Salman H, and Libchaber A. Toward an artificial cell based on gene expression in vesicles. Phys Biol 2005 Sep 15; 2(3) P1-8. doi:10.1088/1478-3975/2/3/P01 pmid:16224117. | ||
Latest revision as of 20:03, 26 October 2007

Cell-Free: Applications
Our Design
To get around the problem of having bacteria in contact with food or medical devices in the applications of Infector Detector and Cell by Date respectively, we aimed to express reporter DNA constructs within a CFS consisting of various E. coli cell extracts (homemade S30, commercial S30 and commercial S30 pT7) in bulk solution. In parallel, we investigated and attempted to optimize conditions for phospholipid vesicle formation. Future work on our project would then involve combining the two, so that protein expression is achieved inside vesicles that have prolonged chassis lifespan.
In vitro Expression in Bulk Solution
Since we are using E. coli plasmid DNA constructs, we chose to use the compatible E. coli cell extract for in-vitro expression. We tried out four options so as to find out the system most suitable for our two applications.
|
|
| ||
|
| ||
Preparation of Homemade S30 Cell Extract Protocol
Preparation of Homemade S12 Cell Extract Protocol
Preparation of Homemade S30 Reaction Mixture
Preparation of Commercially Available S30 Reaction Mixture
Vesicle Formation
For the formation of phospholipid bilayer vesicles, we chose to use the mineral oil method. The protocols are based on Engineering Asymmetric Vesicles by Sophie Pautot, BJ Frisken, and DA Weitz. (2003) 3 and Vincent Noireaux et al. (2005). 4
The mineral oil method consists of three stages, regardless of composition or equipment:
- Preparation of lipid-oil suspension
- Emulsification of vesicle contents
- Bilayer formation through sedimentation
Cell-Free in Infector Detector
For more details on how Infector Detector integrates CFS into its system, go to its Design page.
Cell-Free in Cell by Date
For more details on how Cell by Date utlizes CFS, go to its Design page. For more details on the determination of CFS capabilities and characterizing it, go to its Testing/Validation page.
References
- Kim TW, Keum JW, Oh IS, Choi CY, Park CG, and Kim DM. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J Biotechnol 2006 Dec 1; 126(4) 554-61. doi:10.1016/j.jbiotec.2006.05.014 pmid:16797767.
- [http://www.promega.com/guides/ive_guide/default.htm In vitro protein expression (Promega)]
- Sophie Pautot, B. J. Frisken and D.A. Weitz: Engineering asymmetric vesicles. Proceedings of the National Academy of Sciences 100, p. 10718-10721 (2003).
- Noireaux V, Bar-Ziv R, Godefroy J, Salman H, and Libchaber A. Toward an artificial cell based on gene expression in vesicles. Phys Biol 2005 Sep 15; 2(3) P1-8. doi:10.1088/1478-3975/2/3/P01 pmid:16224117.