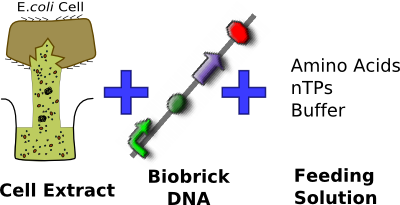Imperial/Cell by Date/Design
From 2007.igem.org
m |
m |
||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:IC07navmenu}} | {{Template:IC07navmenu}} | ||
| + | <br clear="all"> | ||
| + | <html> | ||
| + | <link rel="stylesheet" href="/igem07/index.php?title=User:Dirkvs/Stylesheets/IC07persist.css&action=raw&ctype=text/css" type="text/css" /> | ||
| + | |||
| + | <div id="tabs"> | ||
| + | <ul> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell_by_Date/Introduction" title=""><span>Introduction</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell_by_Date/Specification" title=""><span>Specifications</span></a></li> | ||
| + | <li><a class="current" href="https://2007.igem.org/Imperial/Cell_by_Date/Design" title=""><span>Design</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell_by_Date/Modelling" title=""><span>Modelling</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell_by_Date/Implementation" title=""><span>Implementation</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell_by_Date/Testing" title=""><span>Testing</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell_by_Date/Conclusion" title=""><span>Conclusion</span></a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <hr /> | ||
| + | <br clear="all"> | ||
| + | </html> | ||
__NOTOC__ | __NOTOC__ | ||
| + | = Cell by Date : Design= | ||
| - | = | + | == Design Overview == |
| - | [[Image: | + | [[Image:IC07 design CBD-bb1.png|center||600px]] |
| + | <br> | ||
| - | {| | + | {|border="1" |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| + | |width="100px"|'''Property''' | ||
| + | |width="300px"|'''Value''' | ||
| + | |width="300px"|'''Design Solution''' | ||
| + | |width="100px"|'''System Level''' | ||
| + | |- style="background:#eeffee" | ||
|Health Regulations | |Health Regulations | ||
|System Must not be living replicating bacteria | |System Must not be living replicating bacteria | ||
|Use a Cell Free System e.g. Promega's S30 Cell Extract | |Use a Cell Free System e.g. Promega's S30 Cell Extract | ||
|Chassis | |Chassis | ||
| - | |- | + | |- style="background:#eeffee" |
|Lifespan | |Lifespan | ||
|System must have a shelf life of 7 days | |System must have a shelf life of 7 days | ||
| - | |Protease Inhibitor of Cell Extract | + | |Protease Inhibitor of Cell Extract should ensure degradation of Visual Reporter is Minimal<br> Proper Packaging should ensure that evaporation of Cell Free system is so low that system can surive for 7 days |
|Chassis | |Chassis | ||
| - | |- | + | |- style="background:#eeeeff" |
|Inputs | |Inputs | ||
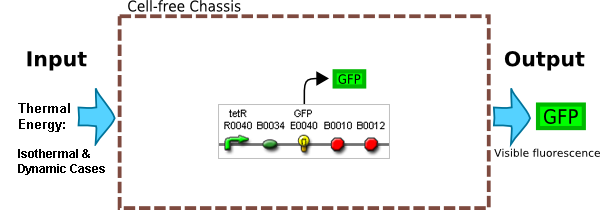
| - | |Isothermal Conditions between 0 & 40 C | + | |Isothermal Conditions between 0 & 40 °C |
|Exploit Thermal Dependance of rates of expression | |Exploit Thermal Dependance of rates of expression | ||
|Construct | |Construct | ||
| - | |- | + | |- style="background:#eeeeff" |
| | | | ||
|Dynamic conditions eg. steps & ramps | |Dynamic conditions eg. steps & ramps | ||
| - | | " | + | |style="text-align:center;"| " |
|Construct | |Construct | ||
| - | |- | + | |- style="background:#eeeeff" |
|Outputs | |Outputs | ||
|System should give a visual signal <br> when beef is off | |System should give a visual signal <br> when beef is off | ||
|Couple constituitive promoter to a Fluoresent Protein eg. RFP | |Couple constituitive promoter to a Fluoresent Protein eg. RFP | ||
|Construct | |Construct | ||
| - | |- | + | |- style="background:#eeeeff" |
|Activation Energy | |Activation Energy | ||
| - | |System Needs to have an activation Energy 30 +/- kJ | + | |System Needs to have an activation Energy 30 +/- kJ mol<sup>-1</sup> |
|To be Determined - this is hard to design for | |To be Determined - this is hard to design for | ||
|Construct | |Construct | ||
| - | |- | + | |- style="background:#eeeeff" |
|Response Time | |Response Time | ||
|System needs to have a response time under 1 hour | |System needs to have a response time under 1 hour | ||
|To be Determined - this is hard to design for | |To be Determined - this is hard to design for | ||
| - | | | + | | Both |
|} | |} | ||
| + | <br clear="all"> | ||
==Chassis Selection== | ==Chassis Selection== | ||
| - | We have chosen to use the commerciall available S30 extract made by Promega. After having looking into a variety of different | + | [[Image:IC07_CFS_components.png|thumb|left|340px|Commercial S30 ''E. coli'' Cell Extract in bulk solution + packaging to last 7 days]] |
| + | |||
| + | We have chosen to use the commerciall available S30 ''E. coli'' cell extract made by Promega. After having looking into a variety of different [[Imperial/Cell-Free/Whatis|cell-free chassis]], we feel that this chassis best suits our needs. In particular this chassis allows us to meet our base requirement of complying with the Health and Safety regulations of the field we are working in, we don't want a live system near our burger meat as potential leak of our system could mean that our system actually spoils the burger meat !! | ||
In addition to complying with health regulations the S30 cell extract is commercially available meaning that it has been shown to work. This is very important for us as it allows our focus to be on tuning the chassis to suit our needs rather than making the chassis work in the first place. | In addition to complying with health regulations the S30 cell extract is commercially available meaning that it has been shown to work. This is very important for us as it allows our focus to be on tuning the chassis to suit our needs rather than making the chassis work in the first place. | ||
| - | ==Construct | + | <br clear="all"> |
| + | |||
| + | |||
| + | ==DNA Constructs== | ||
| + | '''Concept 1'''<br /> | ||
| + | [[Image:IC07 design CBDcon1.png|center||550px]] | ||
| + | <br clear="all"> | ||
| + | |||
| + | |||
| + | '''Concept 2'''<br /> | ||
| + | [[Image:IC07 design CBDcon2.png|center||550px]] | ||
| + | <!-- | ||
| + | {|border=1 width=90% align="center" | ||
| + | |- style="background:#eeeeff" | ||
| + | |width="400px"|'''Construct Design''' | ||
| + | |width="300px"|'''Availability''' | ||
| + | |- | ||
| + | |width="30%"|<center>[[Image:Imperial CBD Design CBD1.png ]]</center> | ||
| + | |width="--"|This part can be found as [http://partsregistry.org/Part:BBa_I13522 BBa_I13522] in the registry. | ||
| + | |- | ||
| + | |<center>[[Image:Imperial CBD Design CBD2.1.png]]</center> | ||
| + | |width="--"|This part can be found as [http://partsregistry.org/Part:BBa_E7104 BBa_E7104] in the Registry, albeit a different RBS and terminator. May require modifications. | ||
| + | |} | ||
| + | <br clear="all"> | ||
| + | --> | ||
| + | In previous iGEM projects, Temperature as an input has been explored through cold shock and heat shock promoters. These promoters essentially only operatre for a limited range of temperatures. | ||
| + | |||
| + | In making a Time Temperature Integrator we would like a promoter that works over a wide range of temperatures, increasing its rate of protein synthesis as temperature increases. We can realise this behaviour by using a simple constituitive promoter and exploiting the thermal dependance of its rate of synthesis, this type of behaviour has been characterised by Ryals.<sup>[[#References |3]]</sup> | ||
| + | |||
| + | In terms of Activation Energy and Response Time we have been unable to find these in literature and to it is hard to make a design that will achieve these targets. However through the course of our experimentation we will determine these properties and hopefullly their values will suit our interests. | ||
| + | |||
| - | |||
| - | + | <center> [https://2007.igem.org/Imperial/Cell_by_Date/Specification << Specifications ] | Design | [https://2007.igem.org/Imperial/Cell_by_Date/Modelling Modelling >>]</center> | |
| - | + | == References == | |
| + | # [http://jb.asm.org/cgi/content/full/180/17/4704?view=long&pmid=9721314 Anne Farewell and Frederick C. Neidhardt. Effect of Temperature on In Vivo Protein Synthetic Capacity in Escherichia coli. J Bacteriol. 1998 September; 180(17): 4704–4710. ] | ||
| + | # [http://www.clemson.edu/clemsonworld/winter2002/6.htm Kent 2002 : Brief about Vitsab TTI] | ||
| + | # [http://jb.asm.org/cgi/reprint/151/2/879?view=long&pmid=6178724 J Ryals, R Little, and H Bremer. Temperature Dependence of RNA Synthesis Parameters in Escherichia coli. J Bacteriol. 1982 August; 151(2): 879–887. ] | ||
Latest revision as of 02:34, 27 October 2007

Cell by Date : Design
Design Overview
| Property | Value | Design Solution | System Level |
| Health Regulations | System Must not be living replicating bacteria | Use a Cell Free System e.g. Promega's S30 Cell Extract | Chassis |
| Lifespan | System must have a shelf life of 7 days | Protease Inhibitor of Cell Extract should ensure degradation of Visual Reporter is Minimal Proper Packaging should ensure that evaporation of Cell Free system is so low that system can surive for 7 days | Chassis |
| Inputs | Isothermal Conditions between 0 & 40 °C | Exploit Thermal Dependance of rates of expression | Construct |
| Dynamic conditions eg. steps & ramps | " | Construct | |
| Outputs | System should give a visual signal when beef is off | Couple constituitive promoter to a Fluoresent Protein eg. RFP | Construct |
| Activation Energy | System Needs to have an activation Energy 30 +/- kJ mol-1 | To be Determined - this is hard to design for | Construct |
| Response Time | System needs to have a response time under 1 hour | To be Determined - this is hard to design for | Both |
Chassis Selection
We have chosen to use the commerciall available S30 E. coli cell extract made by Promega. After having looking into a variety of different cell-free chassis, we feel that this chassis best suits our needs. In particular this chassis allows us to meet our base requirement of complying with the Health and Safety regulations of the field we are working in, we don't want a live system near our burger meat as potential leak of our system could mean that our system actually spoils the burger meat !!
In addition to complying with health regulations the S30 cell extract is commercially available meaning that it has been shown to work. This is very important for us as it allows our focus to be on tuning the chassis to suit our needs rather than making the chassis work in the first place.
DNA Constructs
Concept 1
Concept 2
In previous iGEM projects, Temperature as an input has been explored through cold shock and heat shock promoters. These promoters essentially only operatre for a limited range of temperatures.
In making a Time Temperature Integrator we would like a promoter that works over a wide range of temperatures, increasing its rate of protein synthesis as temperature increases. We can realise this behaviour by using a simple constituitive promoter and exploiting the thermal dependance of its rate of synthesis, this type of behaviour has been characterised by Ryals.3
In terms of Activation Energy and Response Time we have been unable to find these in literature and to it is hard to make a design that will achieve these targets. However through the course of our experimentation we will determine these properties and hopefullly their values will suit our interests.
References
- [http://jb.asm.org/cgi/content/full/180/17/4704?view=long&pmid=9721314 Anne Farewell and Frederick C. Neidhardt. Effect of Temperature on In Vivo Protein Synthetic Capacity in Escherichia coli. J Bacteriol. 1998 September; 180(17): 4704–4710. ]
- [http://www.clemson.edu/clemsonworld/winter2002/6.htm Kent 2002 : Brief about Vitsab TTI]
- [http://jb.asm.org/cgi/reprint/151/2/879?view=long&pmid=6178724 J Ryals, R Little, and H Bremer. Temperature Dependence of RNA Synthesis Parameters in Escherichia coli. J Bacteriol. 1982 August; 151(2): 879–887. ]