Imperial/Cell-Free/Characterisation
From 2007.igem.org
(Difference between revisions)
| Line 8: | Line 8: | ||
| - | + | [[Image:icgems-CBDtemp.JPG|left|600px]] | |
| - | + | <br clear="all"> | |
| Line 15: | Line 15: | ||
| - | + | [[Image:icgems-CBDdnaconc.JPG|left|600px]] | |
| + | <br clear="all"> | ||
<center> [https://2007.igem.org/Imperial/Cell-Free/Contribution << Our Contribution ] | Characterisation | [https://2007.igem.org/Imperial/Cell-Free/Applications Applications >>] | <center> [https://2007.igem.org/Imperial/Cell-Free/Contribution << Our Contribution ] | Characterisation | [https://2007.igem.org/Imperial/Cell-Free/Applications Applications >>] | ||
</center> | </center> | ||
Revision as of 22:17, 20 October 2007

Introduction
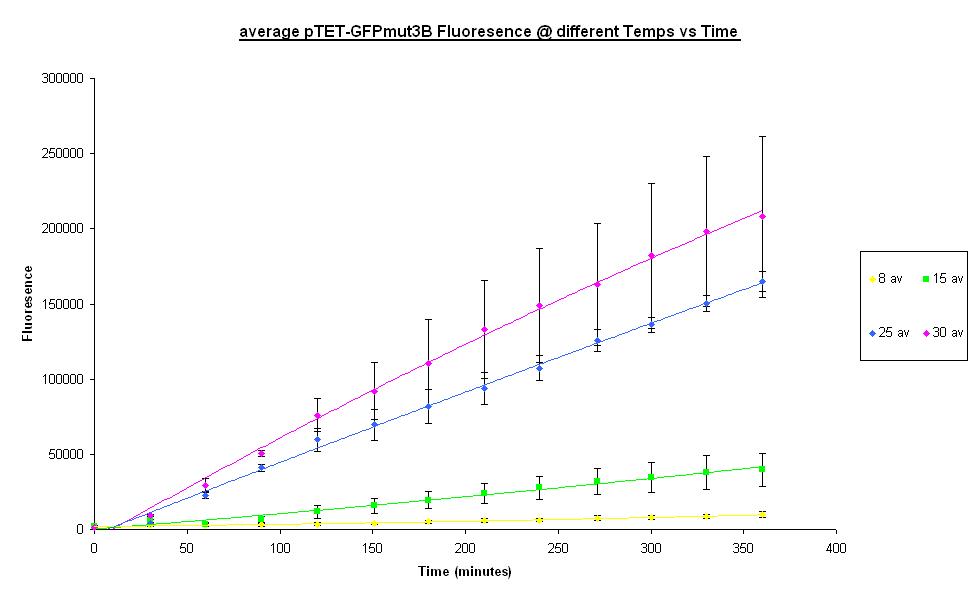
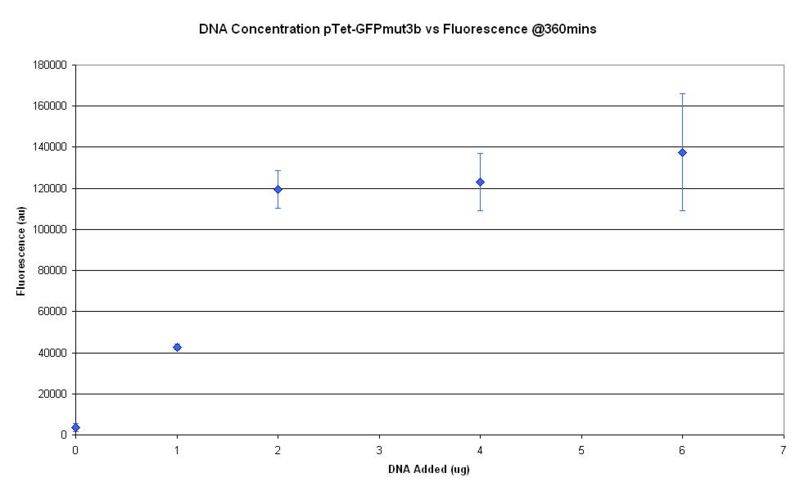
The characterisation of parts and devices in the [http://partsregistry.org/Main_Page Registry of Standard Biological Parts] have have been considered in terms of rate of gene expression in the units Polymerase per Second (PoPS) or Ribosome per Second (RiPS). However the characterisation of the cell-free chassis housing the expression machinery has not been considered up till now. Using [http://www.promega.com Promega's] commercial S30 E. coli cell extract, our team has investigated some parameters that will help us to better understand the characteristics and limitations associated with cell-free systems.
Operating Temperature Ranges
Optimal DNA Concentration