Imperial/Cell-Free/Whatis
From 2007.igem.org
(→References) |
|||
| Line 67: | Line 67: | ||
== References == | == References == | ||
| - | + | # Noireaux V, Bar-Ziv R, and Libchaber A. Principles of cell-free genetic circuit assembly. Proc Natl Acad Sci U S A 2003 Oct 28; 100(22) 12672-7. doi:10.1073/pnas.2135496100 pmid:14559971. | |
| - | + | # Yoshihiro Shimizu, Yutetsu Kuruma, Bei-Wen Ying, So Umekage, Takuya Ueda (2006) Cell-free translation systems for protein engineering FEBS Journal 273 (18), 4133–4140. doi:10.1111/j.1742-4658.2006.05431.x | |
| - | + | # Noireaux V, Bar-Ziv R, Godefroy J, Salman H, and Libchaber A. Toward an artificial cell based on gene expression in vesicles. Phys Biol 2005 Sep 15; 2(3) P1-8. doi:10.1088/1478-3975/2/3/P01 pmid:16224117. | |
<center> On to next stage: | [https://2007.igem.org/Imperial/Cell-Free/Comparison Cell-Free vs. Cell >>]</center> | <center> On to next stage: | [https://2007.igem.org/Imperial/Cell-Free/Comparison Cell-Free vs. Cell >>]</center> | ||
Revision as of 15:28, 23 October 2007

What is Cell-Free?
Cell-free systems (CFS) are based on cell transcription-translation extracts (1), and are essentially non-viable biological systems. Two components define the characteristics of these in vitro translation machineries - the kind of cell extract used, as well as the type of compartmentalization that the cell extract is put in.
Together, cell-free systems can have numerous advantages over in vivo protein synthesis - gene and polymerase concentrations can be controlled, reporter measurements are quantitative, and a large parameter space can be studied (1). This means that CFS could allow the realization of new potential in simple constructs, as well as provide the flexibility to functionalize and establish simplistic controls for the system.
As a forward step to stricter quality control, as well as the specifications of our projects necessitating a cell-free environment, part of our contributions to iGEM involve the investigation and characterization of cell-free expression systems as a new chassis for the Registry.
Types of Extracts
| In vitro synthesis of proteins using cell-free extracts consists of two main processes - transcription of DNA into messenger RNA (mRNA) and translation of mRNA into polypeptides. Coupled transcription-translation systems usually combine a bacteriophage RNA polymerase and promoter (T7, T3, or SP6) with eukaryotic or prokaryotic extracts. In addition, the [http://www.nature.com/nbt/journal/v19/n8/full/nbt0801_732.html PURE] system is a reconstituted CFS for synthesizing proteins using recombinant elements (2). |
Comparison between different types of cell extracts
|
|
|
| |
Types of Compartmentalization
Previous research has been done to optimize cell extracts for in vitro protein synthesis. Their endogenous genetic content is removed so that exogenous DNAs or mRNAs can be expressed. Nuclease activity has been reduced and degradation of certain amino acids has been identified. ATP regenerating systems have also been added to improve the energy supply. Different strategies of compartmentalization have been explored to prolong the lifespan of CFS.

|
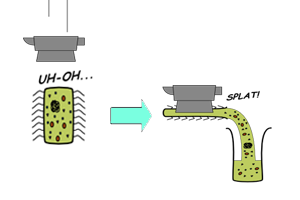
Batch-mode CFS
|
|
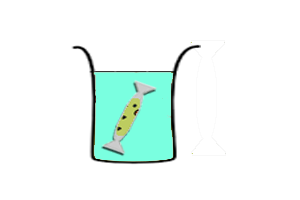
Continuous-exchange CFS
|
|
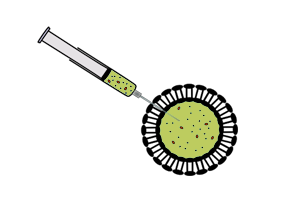
Vesicle-encapsulated CFS
|
References
- Noireaux V, Bar-Ziv R, and Libchaber A. Principles of cell-free genetic circuit assembly. Proc Natl Acad Sci U S A 2003 Oct 28; 100(22) 12672-7. doi:10.1073/pnas.2135496100 pmid:14559971.
- Yoshihiro Shimizu, Yutetsu Kuruma, Bei-Wen Ying, So Umekage, Takuya Ueda (2006) Cell-free translation systems for protein engineering FEBS Journal 273 (18), 4133–4140. doi:10.1111/j.1742-4658.2006.05431.x
- Noireaux V, Bar-Ziv R, Godefroy J, Salman H, and Libchaber A. Toward an artificial cell based on gene expression in vesicles. Phys Biol 2005 Sep 15; 2(3) P1-8. doi:10.1088/1478-3975/2/3/P01 pmid:16224117.