Imperial/Wet Lab/Results/ID3.1
From 2007.igem.org
m |
m |
||
| Line 1: | Line 1: | ||
| + | {{Template: IC07navmenu}} | ||
| + | __NOTOC__ | ||
| + | |||
= ''In vitro'' Testing of pTet-LuxR-pLux-GFPmut3b Construct with varying concentrations of AHL= | = ''In vitro'' Testing of pTet-LuxR-pLux-GFPmut3b Construct with varying concentrations of AHL= | ||
| - | |||
==Aims== | ==Aims== | ||
To characterise the [http://partsregistry.org/Part:BBa_T9002 '''pTet-LuxR-pLux-GFPmut3b'''] construct ''in vitro''. The characterisation is based upon giving inputs of varying AHL concentration and measuring the fluorescent output. | To characterise the [http://partsregistry.org/Part:BBa_T9002 '''pTet-LuxR-pLux-GFPmut3b'''] construct ''in vitro''. The characterisation is based upon giving inputs of varying AHL concentration and measuring the fluorescent output. | ||
Revision as of 12:32, 26 October 2007

In vitro Testing of pTet-LuxR-pLux-GFPmut3b Construct with varying concentrations of AHL
Aims
To characterise the [http://partsregistry.org/Part:BBa_T9002 pTet-LuxR-pLux-GFPmut3b] construct in vitro. The characterisation is based upon giving inputs of varying AHL concentration and measuring the fluorescent output.
Materials and Methods
Link to the Protocols
Results
Controls:
- Negative Control- pTet-LuxR-pLux-GFPmut3b with no AHL
- Negative Control.2.- pLux with 1000nM AHL
Constants:
- Temperature - 25°C
- Volume - 60ul
- Sampling on same plate and every 30 minutes
Raw Data
Discussion
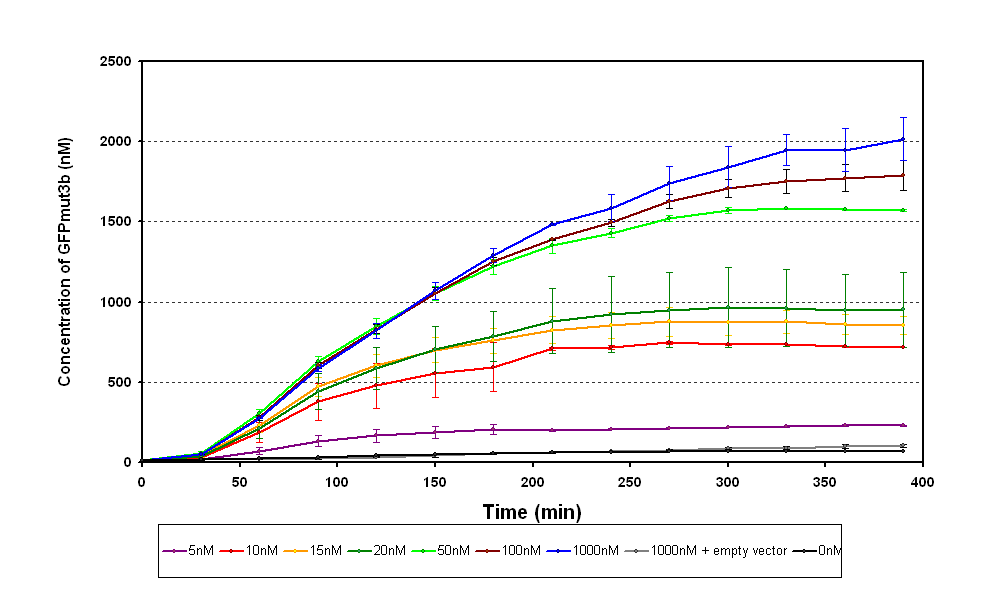
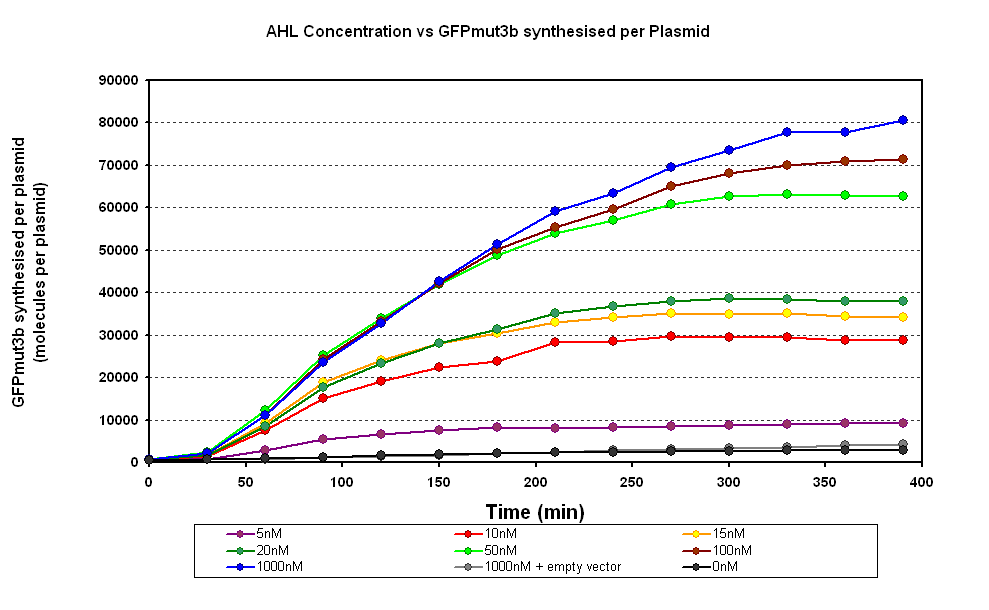
The results from the experiment were of fluorescence against time for varying levels of AHL. Using our calibration curves we managed to convert the fluorescence into concentration of GFPmut3b produced (figure 1.1) and also into Molecules of GFPmut3b synthesised per plasmid (figure 1.2). See the how the calibration curves allowed us to do this by following this link to conversion of units page (LINK).
The idea behind using molecules of GFPmut3b is to try to standardise our units for the chassis and normalise our data based upon the number of DNA plasmids present in our system. This idea is similar to that for the F2620 which uses the units GFP molecules synthesized per second per cell. Trying to normalise this data with the plasmids helps to make our data independent of the DNA used in this assay, i.e. for other constructs tested and characterized in vitro.
There are several key parameters that can be extracted from the data. It looks as if our system becomes saturated to AHL ( the input) above 100nM of AHL, it can be seen that there is little difference between 1000nM and 100nM. If we assume the GFP synthesis 1000nM to be the maximum then the switch point , defined when the output is 50% of the max, is around 20nM of AHL.
The response time is a key characteristic however from this data it is difficult to get an accurate value. The problem is that because the temperature was regulated sampling had to be restricted to every 30 minutes, meaning our best approximation is <30minutes.
The sensitivity of the chassis and construct can also be deduced for the range of AHL concentrations that we looked at. The sensitivity to AHL is <5nM.
Conclusion
These are the characteristics we have extracted
Switch Point= ~20nM AHL
Response Time= <30 minutes
Sensitivity = <5nM AHL
In terms of the specification of the Infector Detector Application the chassis and construct is sensitive enough.