Imperial/Infector Detector/F2620 Comparison
From 2007.igem.org
m (→Summary) |
m |
||
| Line 20: | Line 20: | ||
</html> | </html> | ||
__NOTOC__ | __NOTOC__ | ||
| + | |||
| + | =Summary of Comparison= | ||
| + | The comparison to [http://partsregistry.org/Part:BBa_F2620 F2620] aimed to highlight some of the differences between the chassis and investigate how the constructs characteristics may change between chassis. The [http://partsregistry.org/Part:BBa_F2620 F2620] is an ideal construct to compare for comparison because of its detailed characterisation ''in vivo''. The construct is the same as the construct 1 that was used for infecter detector, '''pTet-LuxR-pLux-GFPmut3b'''. The key findings from the comparison were; | ||
| + | *The normalisation used to compare the two chassis appears to be valid. | ||
| + | *The constructs response appears to be largely independent of the chassis used. | ||
| + | Both of these are important findings and highlight the potential that new chassis offer to synthetic biology. | ||
| + | |||
| + | |||
| - | |||
For further analysis the results of our ''in vitro'' testing have been compared to the work ''in vivo'' on [http://partsregistry.org/Part:BBa_F2620 BBa_F2620]('''pTet-LuxR-pLux-GFPmut3b'''), the construct being the same as our construct 1 for infecter detector. The motivation of the comparison is to see how this construct will respond in different chassis. To do this we investigated a standard unit s to allow the comparison between ''in vitro'' and ''in vivo.'' | For further analysis the results of our ''in vitro'' testing have been compared to the work ''in vivo'' on [http://partsregistry.org/Part:BBa_F2620 BBa_F2620]('''pTet-LuxR-pLux-GFPmut3b'''), the construct being the same as our construct 1 for infecter detector. The motivation of the comparison is to see how this construct will respond in different chassis. To do this we investigated a standard unit s to allow the comparison between ''in vitro'' and ''in vivo.'' | ||
Revision as of 00:42, 26 October 2007

Summary of Comparison
The comparison to [http://partsregistry.org/Part:BBa_F2620 F2620] aimed to highlight some of the differences between the chassis and investigate how the constructs characteristics may change between chassis. The [http://partsregistry.org/Part:BBa_F2620 F2620] is an ideal construct to compare for comparison because of its detailed characterisation in vivo. The construct is the same as the construct 1 that was used for infecter detector, pTet-LuxR-pLux-GFPmut3b. The key findings from the comparison were;
- The normalisation used to compare the two chassis appears to be valid.
- The constructs response appears to be largely independent of the chassis used.
Both of these are important findings and highlight the potential that new chassis offer to synthetic biology.
For further analysis the results of our in vitro testing have been compared to the work in vivo on [http://partsregistry.org/Part:BBa_F2620 BBa_F2620](pTet-LuxR-pLux-GFPmut3b), the construct being the same as our construct 1 for infecter detector. The motivation of the comparison is to see how this construct will respond in different chassis. To do this we investigated a standard unit s to allow the comparison between in vitro and in vivo.
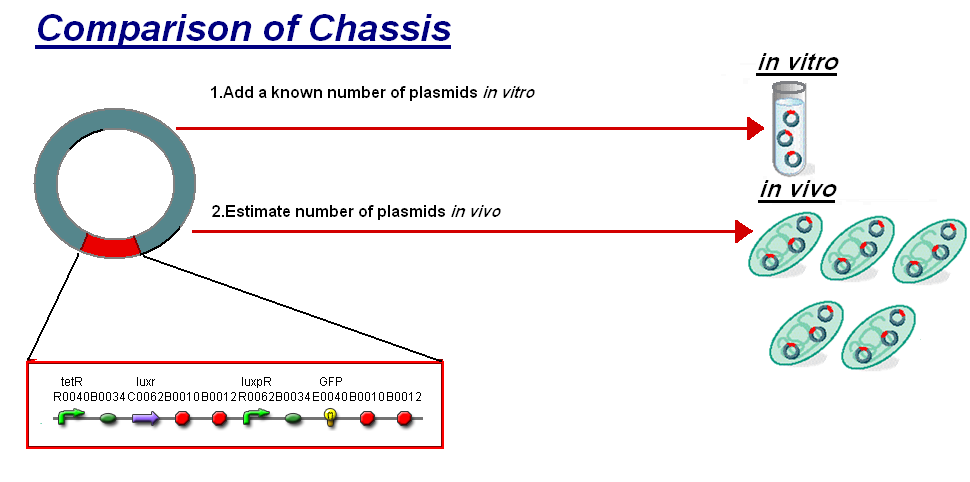
The basis for comparison is to normalise the in vitro chassis on the number of plasmids to give a platform for comparison:
- In Vitro - 4µg of DNA was added which for [http://partsregistry.org/Part:BBa_T9002 pTet-LuxR-pLux-GFPmut3b] is 904823007 plasmids
- In Vivo - Each cell the plasmids number was estimated at 30 per cell
To compare we normalised the data of in vitro GFPmut3b molecules synthesised per 30 plasmids to allow some comparison to the in vivo data.
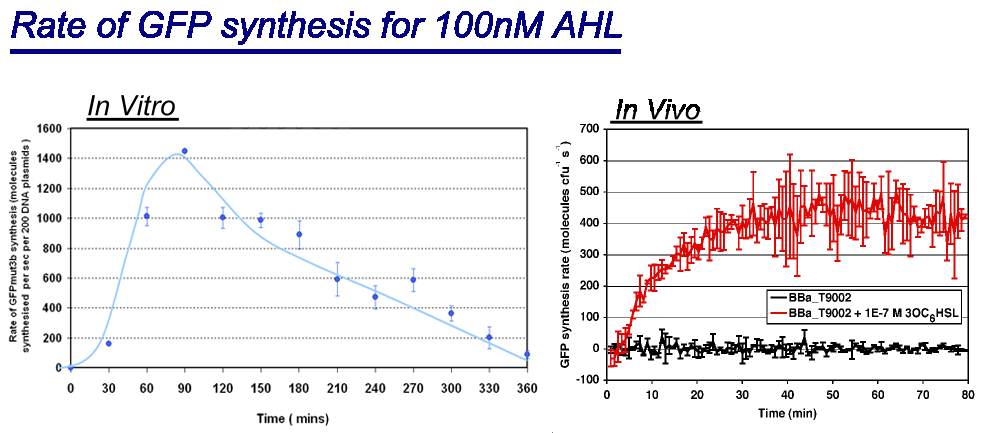
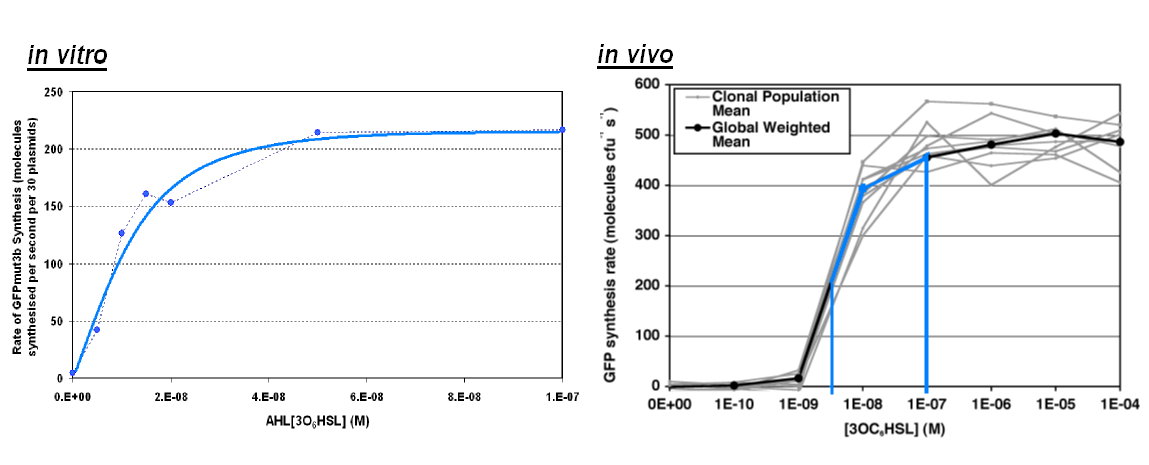
Of particular interest was to compare the:
- Rate of GFP synthesis of 100nM
- Transfer Function.