Imperial/Cell by Date/Testing
From 2007.igem.org
m (→Summary) |
m (→Results) |
||
| Line 52: | Line 52: | ||
[[Image:CBD Arrhenius Plot.PNG|centre|thumb|600px|Fig 2: Arrhenius Plot]] <br clear = "all" > | [[Image:CBD Arrhenius Plot.PNG|centre|thumb|600px|Fig 2: Arrhenius Plot]] <br clear = "all" > | ||
| - | |||
| - | |||
| - | |||
| - | |||
'''Other practical considerations:''' <br> | '''Other practical considerations:''' <br> | ||
*Unfortunetly we have not been able to use a visible reporter and so have no idea whether the construct can in fact produce a visible signal. <br> | *Unfortunetly we have not been able to use a visible reporter and so have no idea whether the construct can in fact produce a visible signal. <br> | ||
*In terms of lifespan we have had major problems with evaporation meaning that the lifespan of our system is limited to a few days as discussed in our packaging experiment below. <br> | *In terms of lifespan we have had major problems with evaporation meaning that the lifespan of our system is limited to a few days as discussed in our packaging experiment below. <br> | ||
| + | |||
| + | ===<font color=blue>Investigating properties of system under dynamic temperature scenarios</font>=== | ||
| + | |||
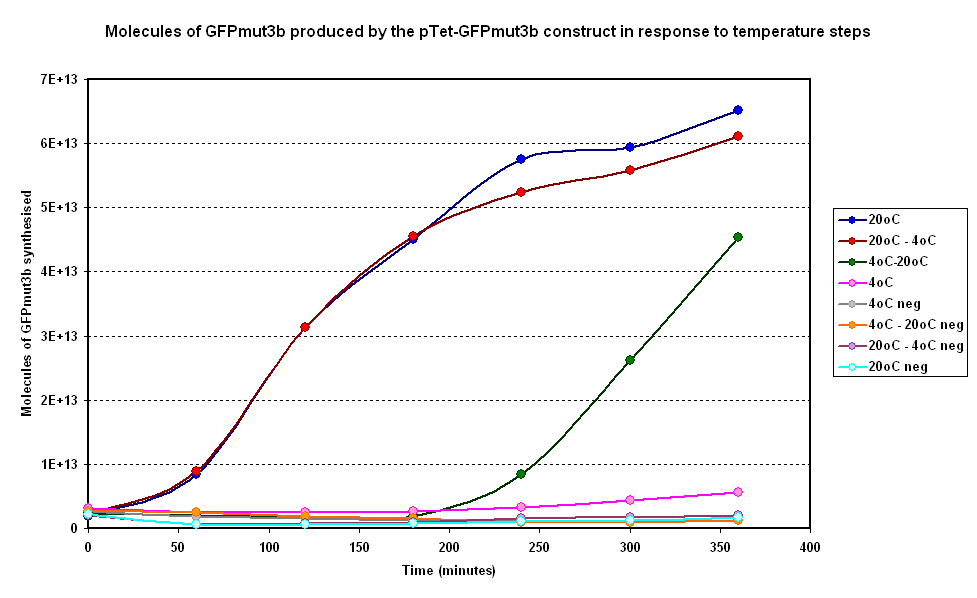
| + | [[Image:IC 2007 CBD steps.PNG|600px|center|thumb|Fig 3:Moelcules of GFPmut3b produced on average over time for two samples at constant temperatures of 4°C and 20°C and two with a temperature step (4-20°C and 20-4°C)]] <br clear = "all"> | ||
| + | |||
| + | qualitative analysis: | ||
| + | |||
| + | 20-4 slow down as per meat model -> Good sign | ||
| + | |||
| + | 4-20 gets going quickly as per meat -> Good sign | ||
| + | |||
| + | Hard to quantify response time due to lack of data points would like to look into this | ||
| + | |||
| + | |||
===<font color=blue>Other Experiments</font>=== | ===<font color=blue>Other Experiments</font>=== | ||
Revision as of 01:20, 27 October 2007

Cell by Date: Testing
Summary
Key results of our experiments are :
1.Operating Range : Our System Operates between 8-37 Degrees
2.Activation Energy : The activation energy of our system is 1.5kJ/mol
3.Optimum DNA Concentration : The optimum DNA concentration for our system is 2-4ug DNA
Aims
Having determined that our system worked at a basic level in our implementation phase we set out to ascertain a clearer picture of the performance of our system to ultimately reveal whether our design had realised our specifications.
Results
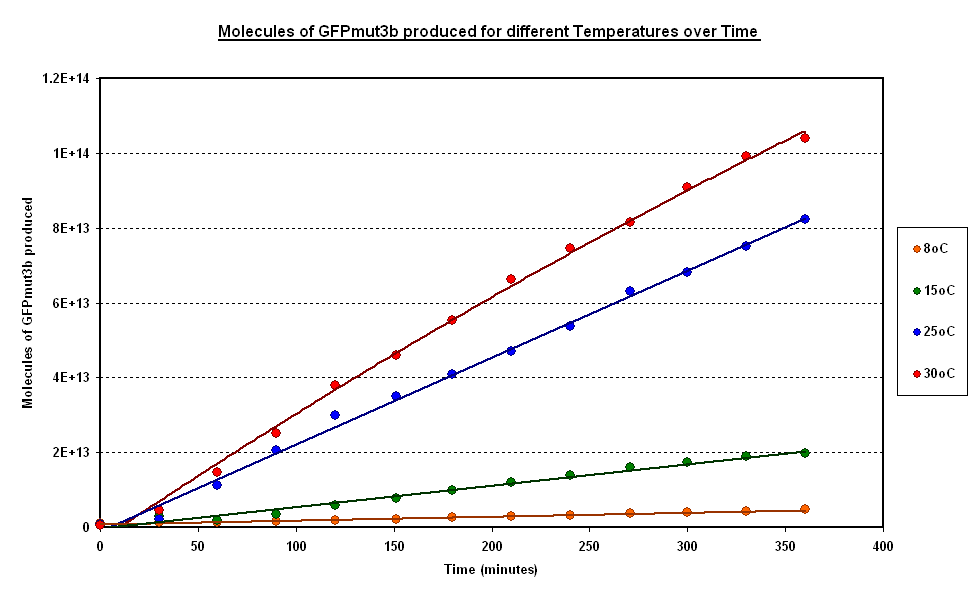
Investigating properties of system under isothermal scenarios
To estimate the properties of our system we looked at the evolution of the Fluorescence with time for experiments where temperature is carefully kept constant. (Figure 1).
General Behaviour:
- In all experiment we can observe a linear growth of fluorescence corresponding to a steady rate of protein production.
- This matches the claims of the manufacturer of the cell-free extract that the extract is optimised so that protein degradation is negligible.
Operating Range:
- Our system seems to 'turn off' at around 4 degrees C.
- At 8 degrees with have minimal expression which then increases to a substantial expression at 37 degrees.
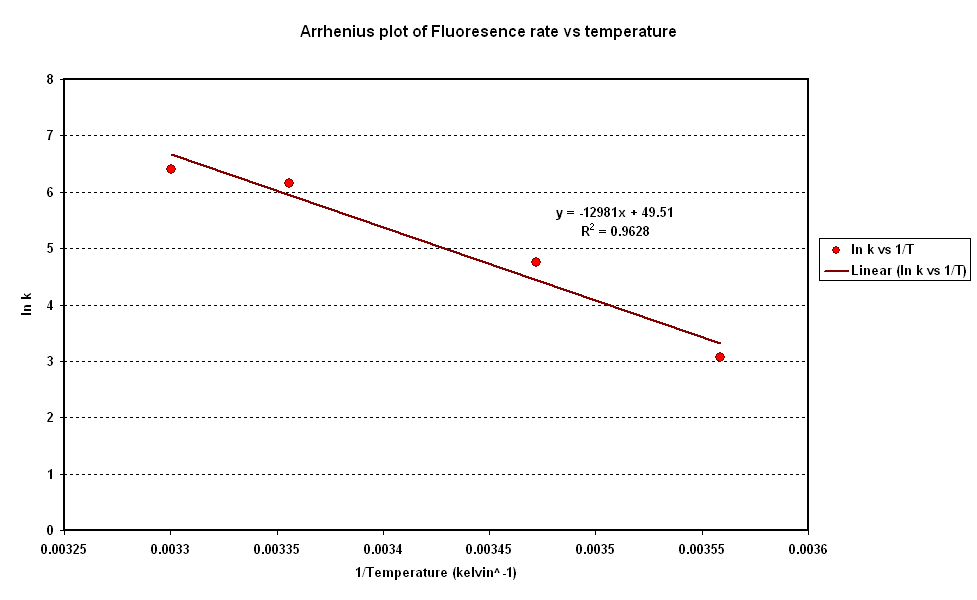
Synthesis rate vs Temperature
- Over the range of temperature considered here the rate increases with temperature.
- To investigate the relation between synthesis rate and temperature we extracted the rate of synthesis of GFP for each experiment and plotted it against 1/T (Figure 2).
- A strong linear correlation in the log plot supports an arrhenius type dependence on temperature with an activation energy of 1.5kJ/mol.
Other practical considerations:
- Unfortunetly we have not been able to use a visible reporter and so have no idea whether the construct can in fact produce a visible signal.
- In terms of lifespan we have had major problems with evaporation meaning that the lifespan of our system is limited to a few days as discussed in our packaging experiment below.
Investigating properties of system under dynamic temperature scenarios
qualitative analysis:
20-4 slow down as per meat model -> Good sign
4-20 gets going quickly as per meat -> Good sign
Hard to quantify response time due to lack of data points would like to look into this
Other Experiments
1. DNA Concentration
2. Packaging