Imperial/Cell-Free/Characterisation
From 2007.igem.org
| Line 3: | Line 3: | ||
== Cell-Free: Characterisation == | == Cell-Free: Characterisation == | ||
| - | The characterisation of parts and devices in the [http://partsregistry.org/Main_Page Registry of Standard Biological Parts] have have been considered in terms of rate of gene expression in the units Polymerase per Second (PoPS) or Ribosome per Second (RiPS). However the characterisation of the cell-free chassis housing the expression machinery has not been considered up till now. Using [http://www.promega.com Promega's] commercial S30 ''E. coli'' cell extract, our team has investigated some parameters that will help us | + | The characterisation of parts and devices in the [http://partsregistry.org/Main_Page Registry of Standard Biological Parts] have have been considered in terms of rate of gene expression in the units Polymerase per Second (PoPS) or Ribosome per Second (RiPS). However the characterisation of the cell-free chassis housing the expression machinery has not been considered up till now. Using [http://www.promega.com Promega's] commercial S30 ''E. coli'' cell extract in batch mode, our team has investigated some parameters that will help us better understand the characteristics and limitations associated with cell-free systems. |
| - | === Operating Temperature | + | === Operating Temperature Range === |
| - | + | {| style="width:800px;" cellpadding="2" cellspacing="0" align="center" | |
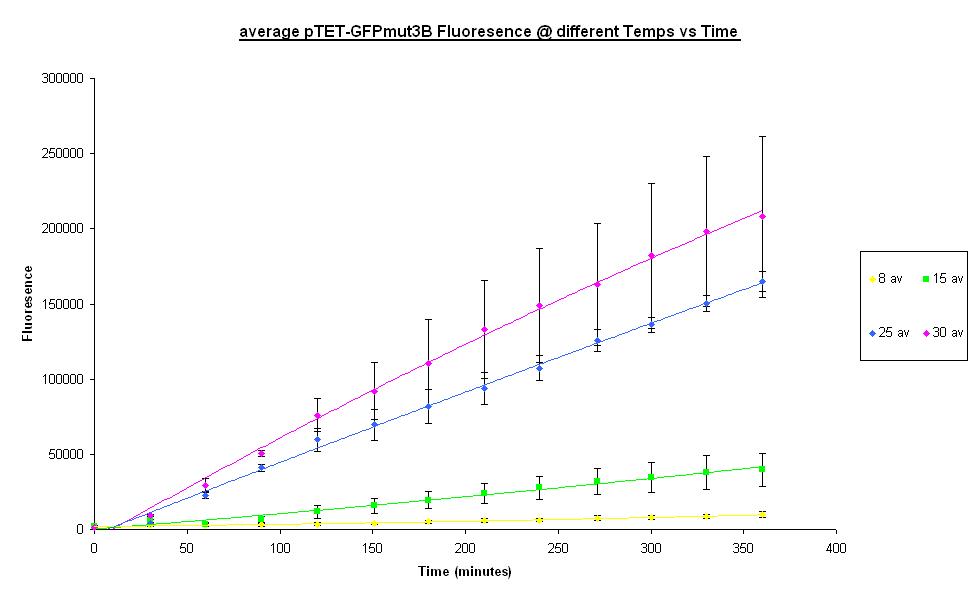
| - | [[Image:icgems-CBDtemp.JPG|left| | + | |- style="background:#ffffff; text-align:left;" color:white;" align="center" |
| + | ||[[Image:icgems-CBDtemp.JPG|left|500px]] | ||
| + | |style="padding-left:30px;"| Insert raw data and results writeup here for 8, 15, 25, 30, 37, and 45 oC. | ||
| + | |} | ||
<br clear="all"> | <br clear="all"> | ||
| + | The characterization of various expression levels at different temperatures is achieved using experiments for Cell by Date. As shown, there is a positive correlation of levels of fluorescence and hence expression that the system produces as temperature increases from 8°C through 15°C, 25°C to 30°C. Unfortunately, he limits of the operating temperature range has not been characterized. It is however known from the other experiments conducted that the system operates at 37°C, but not at 45°C. | ||
=== Optimal DNA Concentration === | === Optimal DNA Concentration === | ||
| + | {| style="width:800px;" cellpadding="2" cellspacing="0" align="center" | ||
| + | |- style="background:#ffffff; text-align:left;" color:white;" align="center" | ||
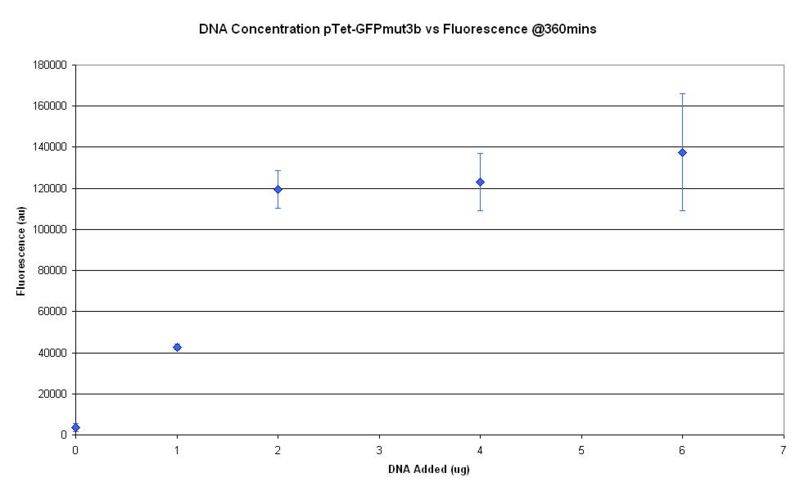
| + | || [[Image:icgems-CBDdnaconc.JPG|left|500px]] | ||
| + | |style="padding-left:30px;"| Insert raw data and results writeup here for optimal DNA Concentration | ||
| + | |} | ||
| + | <br clear="all"> | ||
| + | One of the advantages of using CFS is better controllability of DNA in the system. While the introduction of DNA is easily achieved by its addition into the batch solution, optimal DNA concentration in the CFS needs to be ascertained. | ||
| - | + | Our results show that the difference in DNA addition of more than 2μg into a 60ml batch solution... doesnt mean a thing! | |
| - | + | ||
<center> [https://2007.igem.org/Imperial/Cell-Free/Contribution << Our Contribution ] | Characterisation | [https://2007.igem.org/Imperial/Cell-Free/Applications Applications >>] | <center> [https://2007.igem.org/Imperial/Cell-Free/Contribution << Our Contribution ] | Characterisation | [https://2007.igem.org/Imperial/Cell-Free/Applications Applications >>] | ||
</center> | </center> | ||
Revision as of 23:07, 20 October 2007

Cell-Free: Characterisation
The characterisation of parts and devices in the [http://partsregistry.org/Main_Page Registry of Standard Biological Parts] have have been considered in terms of rate of gene expression in the units Polymerase per Second (PoPS) or Ribosome per Second (RiPS). However the characterisation of the cell-free chassis housing the expression machinery has not been considered up till now. Using [http://www.promega.com Promega's] commercial S30 E. coli cell extract in batch mode, our team has investigated some parameters that will help us better understand the characteristics and limitations associated with cell-free systems.
Operating Temperature Range
| Insert raw data and results writeup here for 8, 15, 25, 30, 37, and 45 oC. |
The characterization of various expression levels at different temperatures is achieved using experiments for Cell by Date. As shown, there is a positive correlation of levels of fluorescence and hence expression that the system produces as temperature increases from 8°C through 15°C, 25°C to 30°C. Unfortunately, he limits of the operating temperature range has not been characterized. It is however known from the other experiments conducted that the system operates at 37°C, but not at 45°C.
Optimal DNA Concentration
| Insert raw data and results writeup here for optimal DNA Concentration |
One of the advantages of using CFS is better controllability of DNA in the system. While the introduction of DNA is easily achieved by its addition into the batch solution, optimal DNA concentration in the CFS needs to be ascertained.
Our results show that the difference in DNA addition of more than 2μg into a 60ml batch solution... doesnt mean a thing!