Imperial/Infector Detector/Testing
From 2007.igem.org
(→Summary) |
m |
||
| Line 12: | Line 12: | ||
<li><a href="https://2007.igem.org/Imperial/Infector_Detector/Implementation" title=""><span>Implementation</span></a></li> | <li><a href="https://2007.igem.org/Imperial/Infector_Detector/Implementation" title=""><span>Implementation</span></a></li> | ||
<li><a class="current" href="https://2007.igem.org/Imperial/Infector_Detector/Testing" title=""><span>Testing</span></a></li> | <li><a class="current" href="https://2007.igem.org/Imperial/Infector_Detector/Testing" title=""><span>Testing</span></a></li> | ||
| + | <li><a class="current" href="https://2007.igem.org/Imperial/Infector_Detector/F2620 Comparison" title=""><span>Testing</span></a></li> | ||
<li><a href="https://2007.igem.org/Imperial/Infector_Detector/Conclusion" title=""><span>Conclusion</span></a></li> | <li><a href="https://2007.igem.org/Imperial/Infector_Detector/Conclusion" title=""><span>Conclusion</span></a></li> | ||
</ul> | </ul> | ||
Revision as of 22:49, 25 October 2007

Infector Detector: Testing
Aims of Testing
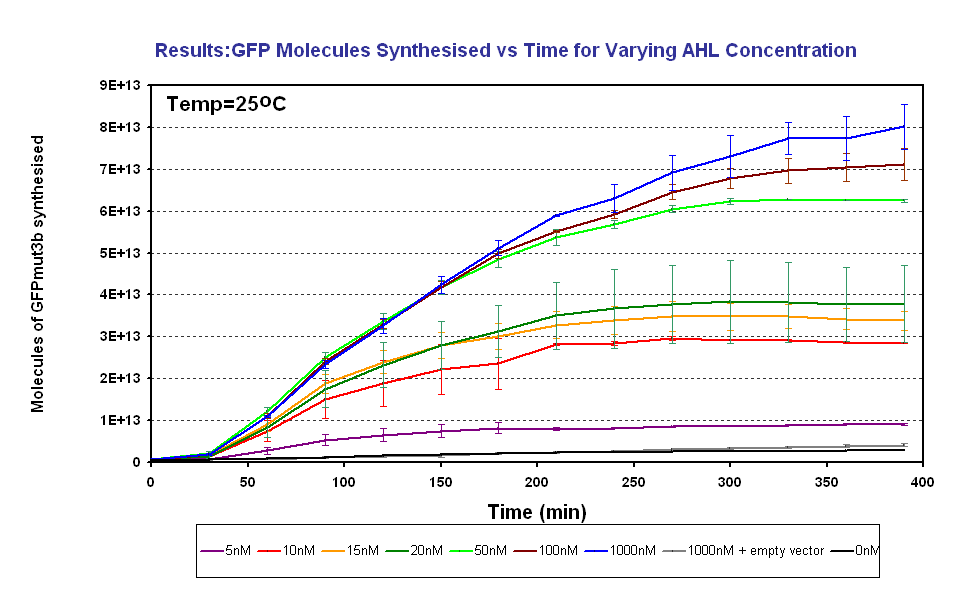
To test and characterise the key characteristics of our system such as the sensitivity of the system to AHL. To do this, we induce the system with known concentrations of AHL input and measure the fluorescence output. Then using a calibration curve the fluorescence was converted into the number of GFPmut3b molecules synthesised, click on the following link for an explaination about how to [http://www.openwetware.org/wiki/IGEM:IMPERIAL/2007/new_pages/Data_anaylsis#Conversion_of_Datause use the calibration curve]. The full results and protocols can be found on the links [http://openwetware.org/wiki/IGEM:Imperial/2007/Wet_Lab/results/ID3.1 results] and protocol pages.
Results
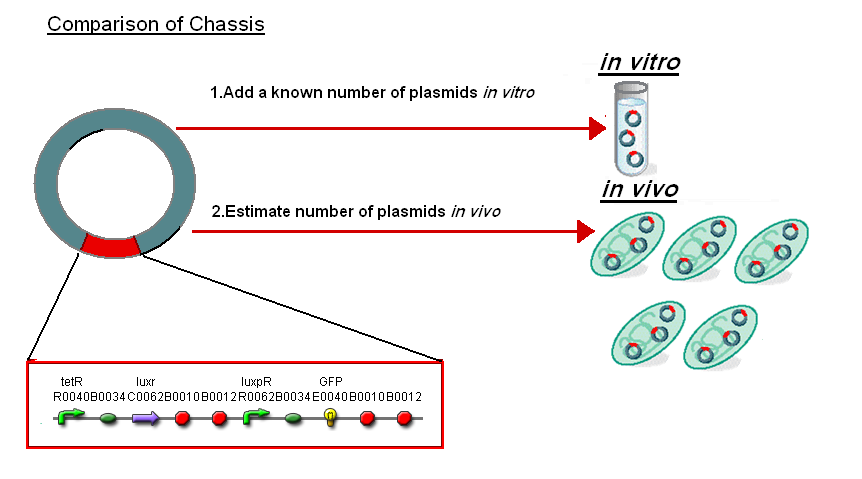
Normalise the in vitro on the plasmids to give a platform for comparison:
- In Vitro - 4µg of DNA was added which for [http://partsregistry.org/Part:BBa_T9002 pTet-LuxR-pLux-GFPmut3b] is 904823007 plasmids
- In Vivo - Each cell has ~30 plasmids per cell
To compare we normalised the data of in vitro GFPmut3b molecules synthesised per 30 plasmids to allow some comparison to the in vivo data.
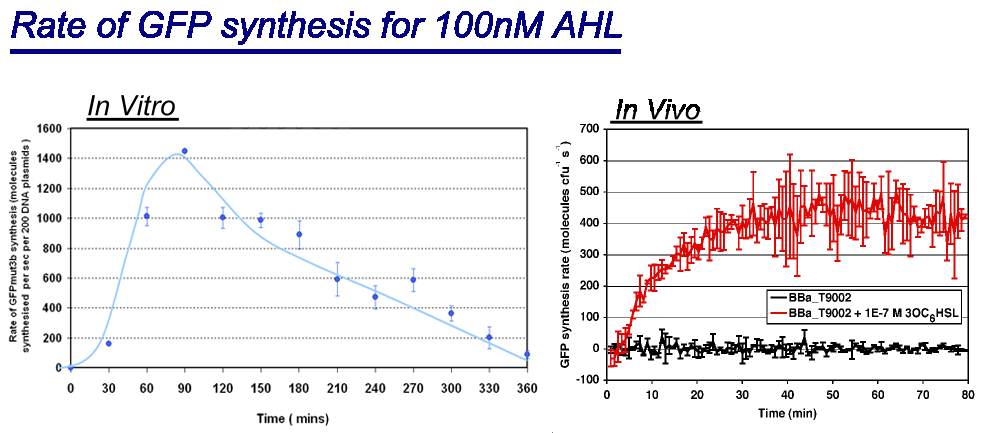
Rate of GFPmut3b Synthesis for 100nM AHL
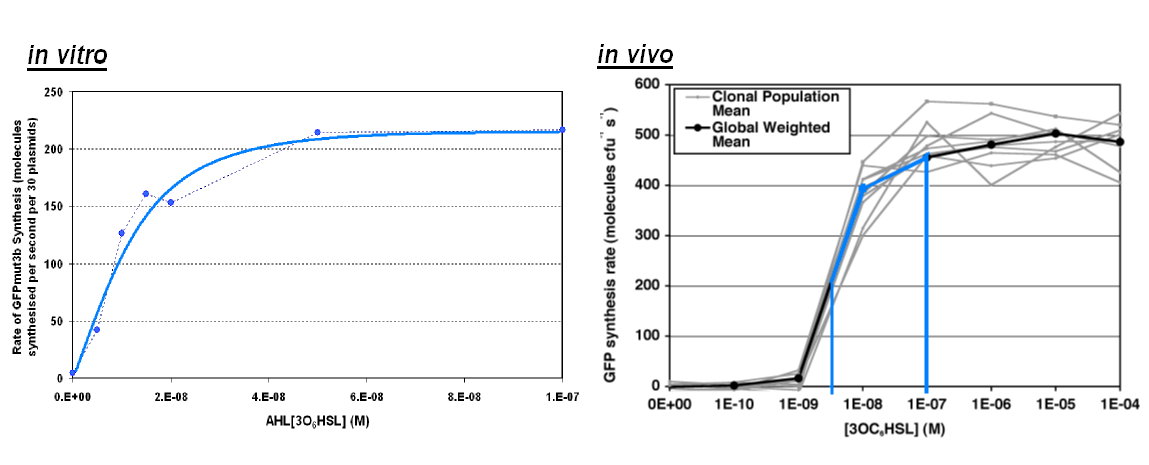
Transfer Function
Summary
Below is list of which of the orginial Specifications that our infecter detector achieved:
| Achievements | ||
| Inputs | Sensitive to 5-1000nM | |
| Outputs | Future work - Using Stronger fluorescent protein such as DsRed express | |
| Response Time | Systems responds <30minutes | |
| Operating Conditions | System works at 25°C | |
| Health & Safety | Cell Free in vitro chassis | |
| Lifespan | Can be stored in freezer for prolonged periods | |
| Packaging | Future Work - Using our chassis in a spray |