Imperial/Infector Detector/F2620 Comparison
From 2007.igem.org
m (→Transfer Function) |
(→Summary of Comparison) |
||
| Line 22: | Line 22: | ||
=Summary of Comparison= | =Summary of Comparison= | ||
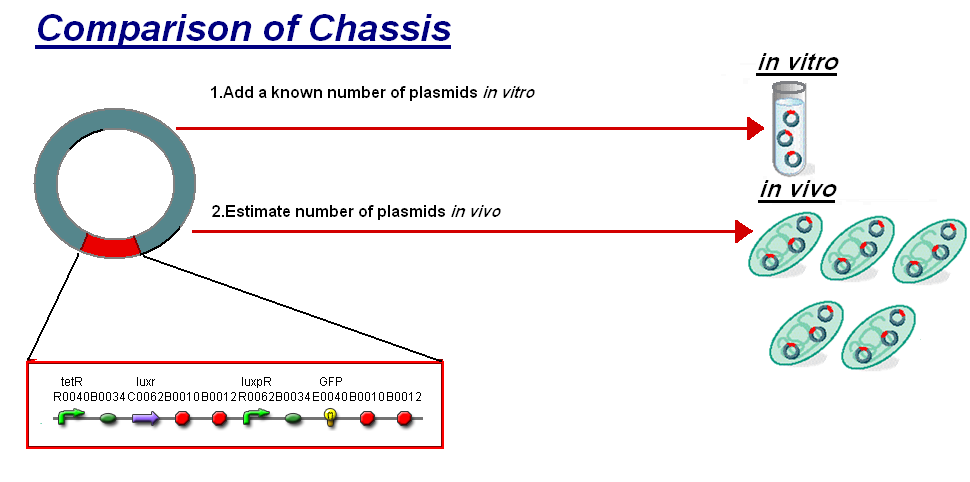
| - | We thought to compare our ''in vitro'' characterisation to the characterisation of [http://partsregistry.org/Part:BBa_F2620 F2620] ''in vivo'' with the aim to highlight some of the differences between the chassis and investigate how the | + | We thought to compare our ''in vitro'' characterisation to the characterisation of [http://partsregistry.org/Part:BBa_F2620 F2620] ''in vivo'' with the aim to highlight some of the differences between the chassis and investigate how the construct's characteristics may change between them. The [http://partsregistry.org/Part:BBa_F2620 F2620] is an ideal construct for comparison because of its detailed characterisation ''in vivo''. The construct is the same as the construct 1 that was used for infecter detector, '''pTet-LuxR-pLux-GFPmut3b'''. The key results of the comparison were; |
*The '''creation of a new unit''' to allow comparison between ''in vitro'' and ''in vivo'' chassis. | *The '''creation of a new unit''' to allow comparison between ''in vitro'' and ''in vivo'' chassis. | ||
| - | *That although we are changing the ''E.coli'' chassis from ''in vivo'' to ''in vitro'' the construct characteristic '''response is independent of the chassis'''. | + | *That although we are changing the ''E.coli'' chassis from ''in vivo'' to ''in vitro'' the construct's characteristic '''response is independent of the chassis'''. |
| - | These are exciting findings, revealing the potential for the exploration of new chassis and the ability to use constructs in an exchangable | + | These are exciting findings, revealing the potential for the exploration of new chassis and the ability to use constructs in an exchangable manner. |
Revision as of 21:51, 26 October 2007

Summary of Comparison
We thought to compare our in vitro characterisation to the characterisation of [http://partsregistry.org/Part:BBa_F2620 F2620] in vivo with the aim to highlight some of the differences between the chassis and investigate how the construct's characteristics may change between them. The [http://partsregistry.org/Part:BBa_F2620 F2620] is an ideal construct for comparison because of its detailed characterisation in vivo. The construct is the same as the construct 1 that was used for infecter detector, pTet-LuxR-pLux-GFPmut3b. The key results of the comparison were;
- The creation of a new unit to allow comparison between in vitro and in vivo chassis.
- That although we are changing the E.coli chassis from in vivo to in vitro the construct's characteristic response is independent of the chassis.
These are exciting findings, revealing the potential for the exploration of new chassis and the ability to use constructs in an exchangable manner.
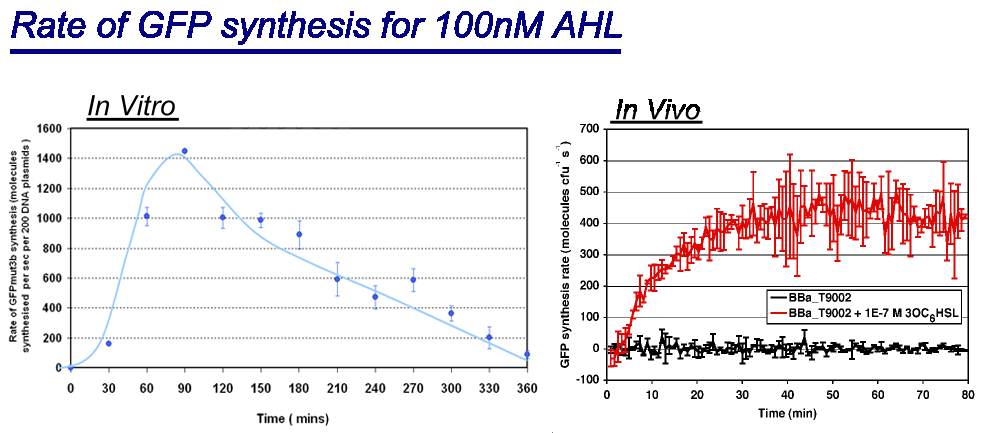 |
| Comparison between in vivo and in vitro for rate of GFPmut3b synthesis for 100nM AHL. The in vivo chassis used was the bacterial strain MG1655 and the in vitro chassis was Promega Commercial S30 Cell Extract(60µl)
Key Difference:
Interestingly the values of rate of synthesis are in the same order magnitude of hundreds, this suggesting that the normalisation we are using to compare these chassis is valid. |
Transfer Function
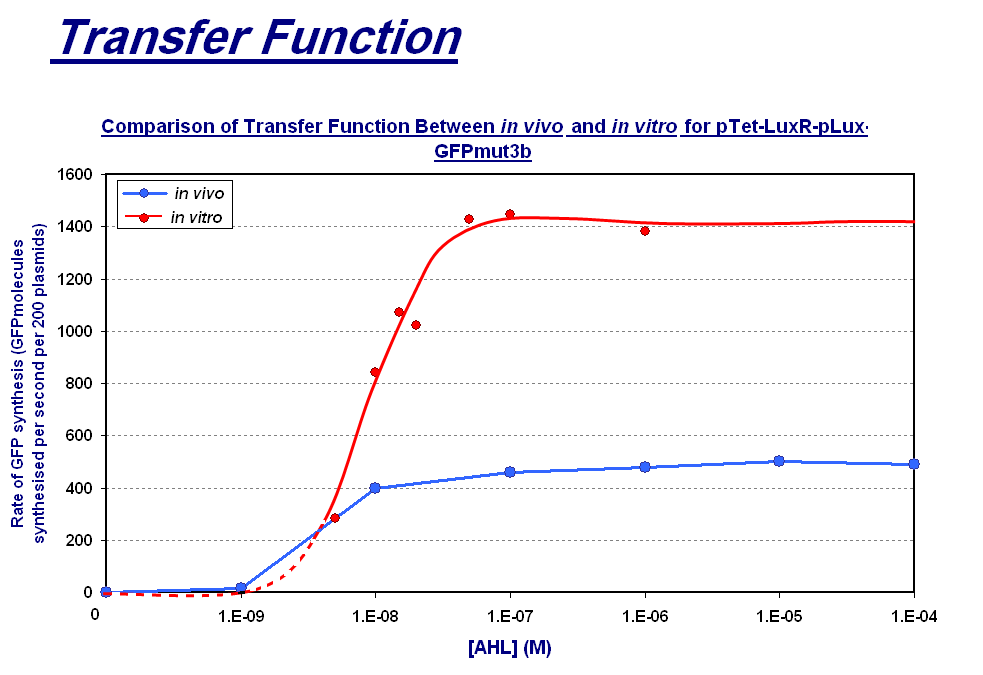 |
| The graph above shows the transfer function of [AHL] input vs rate of GFP synthesis output. The graph shows the max rate of synthesis for each of the chassis; for in vivo this is the steady state reached after about 30 minutes and for in vitro it is the rate between 60 and 90 minutes which is the maximum rate before the energy limitations of the system cause the rate to drop. The blue line on corresponds to the range of AHL and the response of the in vitro chassis.
|