Imperial/Wet Lab/Protocols/CBD1.1
From 2007.igem.org

Wet Lab: Protocols: Initial Testing of DNA Constructs in vivo
Aims
- To test to see if the DNA constructs from the registry are viable. This is done in vivo.
- The constructs are pTet-GFP and pT7-GFP.
Day 1
Equipment
- 7ml sterile tubes x4
- 1.5ml Eppendorf tube x1
- 37°C incubator
- Gilson Pipettes p1000 p200 p20
Reagents
- E.coli BL21; culture containing parts :pTet-GFP, pT7-GFP, pcI-GFP
- LB medium
- Ampicillin stock (50 mg/ml)
Protocol
Innoculation of Media
- Inoculate 10ul of transformed E.coli cells in individual 2ml LB medium containing 2ul of ampicillin
- Incubate at 37°C for overnight in a shaker. (This is to get a good stock of cells for use in the experiment. After the overnight culture the cells will be in stationary phase)
Day 2
Equipment
- 1 Fluorometer plate (black)
- Fluorometer + PC
- Gilson pipettes 1000 and 200
- Eppendorf tubes
Reagents
- LB medium
- E.coli culture with transformed plasmid
- GFP standard solution
- ddH2O
Protocol
Preparation of diluted GFP standard solution
- Add 995ul of ultra pure water an eppendorf tube, together with 5ul of undiluted GFP standard solution and mix. (This gives a 200x dilution to be used as a positive control)
- Place the tube on ice till it is ready to be used.
Loading Plate
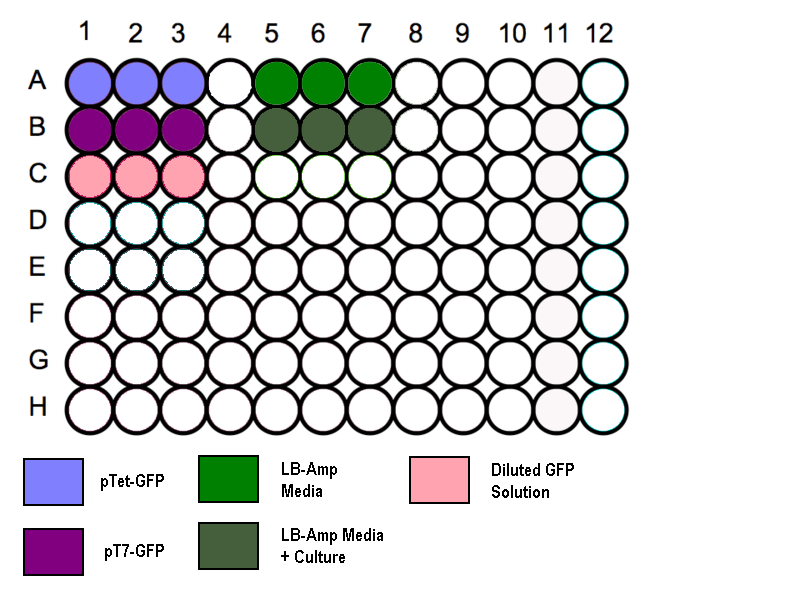
- Transfer 200 µl aliquots of each of the cultures to a flat-bottomed 96 well plate. (Follow the schematic as shown)
- Three wells to be filled with 200µl of media to measure the absorbance background.
- Standard GFP solution added as a positive control.
- Remove lid and measure in the flourometer.
- (Fluorescence measurements - 488 nm excitation filter, 525 nm emission filter, 0.5 seconds, CW lamp energy 12901 units)
- Repeat the measurement a further two times straight after each other (This is to test the variability of the machine)
Schematic
|