Imperial/Infector Detector/Testing
From 2007.igem.org

Infector Detector: Testing
Aims
The aims of the testing were as follows:
- To test and obtain the optimal DNA concentration for construct 1 in vitro
- To characterise the output of GFPmut3b for a range of AHL inputs. From this obtain the AHL sensitivity of our system.
In addition the fluorescence measurements were converted to number of GFPmut3b molecules synthesised using a calibration curve constructed using purified GFPmut3b.
Results
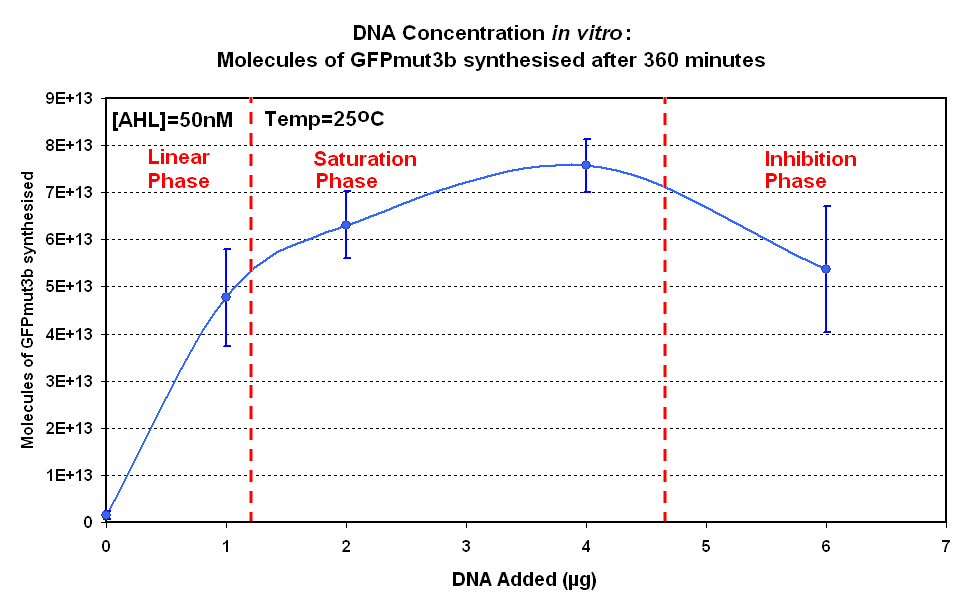
DNA Concentrations
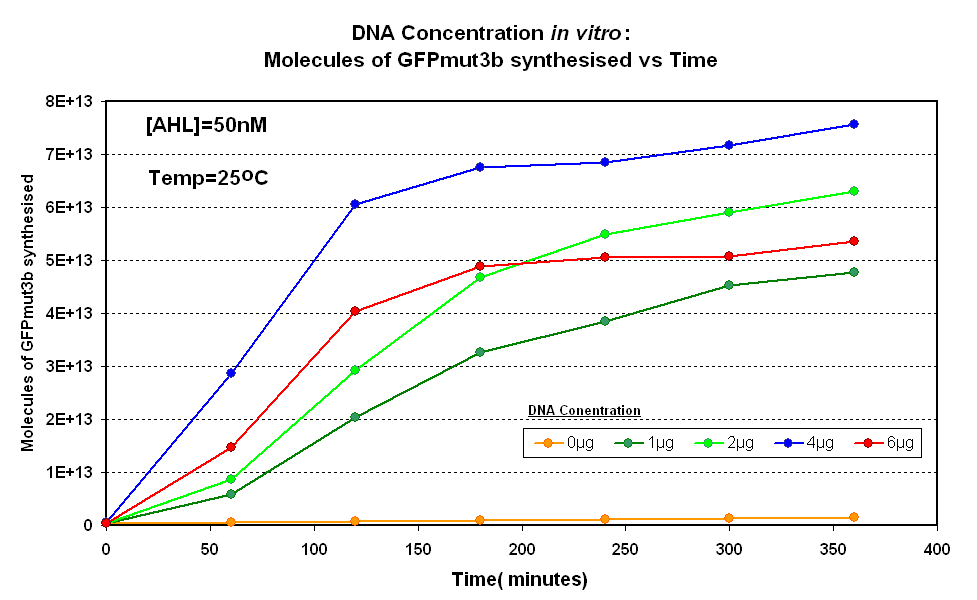 Fig.1.1:Molecules of GFPmut3b synthesised over time, for each DNA Concentration in vitro - The fluorescence was measured over time for each experiment and converted into molecules of GFPmut3b in vitro using our calibration curve. |
The Results above show that the optimum DNA concentration for in vitro is 4µg. From figure 1.1. and 1.2 it can be seen that as DNA concentration increases above 4µg the GFPmut3b molecules synthesised decrease. Interestingly for figure 1.2 the graph can be split into several regions of how the DNA concentration changes the GFPmut3b synthesis. Between
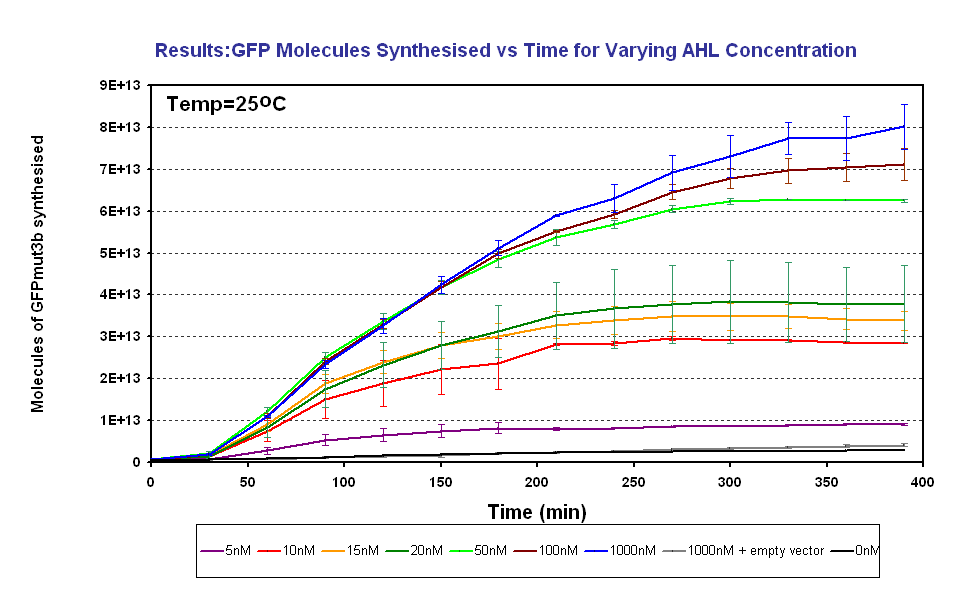
Testing AHL Range
The results show us the following:
|